A Comprehensive Guide to the Morphological Features of Apoptosis Phases I, IIa, and IIb
This article provides a detailed examination of the distinct morphological characteristics that define Phase I (early), Phase IIa (middle), and Phase IIb (late) of apoptosis.
A Comprehensive Guide to the Morphological Features of Apoptosis Phases I, IIa, and IIb
Abstract
This article provides a detailed examination of the distinct morphological characteristics that define Phase I (early), Phase IIa (middle), and Phase IIb (late) of apoptosis. Tailored for researchers, scientists, and drug development professionals, it bridges foundational knowledge with practical application. The content systematically explores the ultrastructural changes observed via various microscopy techniques, compares methodological approaches for detection and analysis, addresses common challenges in morphological identification, and validates findings through integration with biochemical assays. This guide serves as a critical resource for accurately identifying and quantifying apoptotic progression in experimental and clinical contexts, ultimately informing therapeutic development.
Defining the Morphological Hallmarks of Apoptotic Phases I, IIa, and IIb
Apoptosis, a fundamental programmed cell death process, is characterized by a series of distinctive morphological and biochemical hallmarks that enable the controlled elimination of cells without inducing inflammation. This highly regulated process is crucial for multicellular organisms, playing essential roles in embryogenesis, tissue homeostasis, and the removal of damaged or potentially harmful cells [1] [2]. The execution of apoptosis occurs through specific signaling pathways—primarily the intrinsic and extrinsic pathways—that converge on the activation of caspases, which systematically dismantle cellular components [2]. Understanding the precise morphological features of apoptosis and their underlying molecular mechanisms has significant implications for therapeutic interventions in cancer, neurodegenerative disorders, and autoimmune diseases. This technical review provides an in-depth analysis of apoptosis within the context of morphological research, detailing experimental methodologies, key regulatory networks, and essential research tools for investigating this critical cellular process.
Morphological Hallmarks of Apoptosis
The identification of apoptosis relies heavily on recognizing its characteristic morphological features, which distinguish it from other forms of cell death such as necrosis. These morphological changes occur in a coordinated sequence and can be observed through various microscopic techniques.
Core Morphological Characteristics
Table 1: Key Morphological Features of Apoptosis
| Morphological Feature | Description | Detection Methods |
|---|---|---|
| Cell Shrinkage | Reduction in cell volume and cytoplasmic compaction | Light microscopy, electron microscopy |
| Chromatin Condensation | Chromatin aggregation into dense masses beneath nuclear membrane | Nuclear staining (DAPI, Hoechst), electron microscopy |
| Nuclear Fragmentation | Nuclear breakdown into discrete fragments (karyorrhexis) | Fluorescence microscopy, TUNEL assay |
| Membrane Blebbing | Formation of bulges on plasma membrane surface | Time-lapse microscopy, electron microscopy |
| Apoptotic Body Formation | Cell fragmentation into membrane-bound vesicles containing organelles | Electron microscopy, fluorescence microscopy |
| Phagocytosis | Engulfment of apoptotic bodies by neighboring cells | Histological analysis, time-lapse imaging |
The morphological process of apoptosis begins with cell shrinkage and chromatin condensation, where the nucleus undergoes pyknosis (condensation) and karyorrhexis (fragmentation) [1] [3]. This is followed by extensive plasma membrane blebbing and the separation of cell fragments into membrane-bound apoptotic bodies in a process called budding [3]. These apoptotic bodies contain intact organelles and are rapidly phagocytosed by macrophages, parenchymal cells, or neoplastic cells, subsequently degrading in phagolysosomes [1] [3]. Critically, this entire process occurs without inducing inflammation, as apoptotic cells do not release their cellular contents into the surrounding environment [3].
Contrasting Apoptosis and Necrosis
Table 2: Comparative Analysis: Apoptosis versus Necrosis
| Characteristic | Apoptosis | Necrosis |
|---|---|---|
| Distribution | Affects individual scattered cells | Affects massive contiguous cell groups |
| Cellular Morphology | Cell shrinkage, cytoplasmic compaction | Cell swelling, organelle disruption |
| Nuclear Changes | Chromatin condensation and margination | Irregular chromatin clumping, karyolysis |
| DNA Fragmentation | Internucleosomal cleavage (DNA ladder) | Random DNA degradation (smear pattern) |
| Membrane Integrity | Maintained until late stages | Lost early in the process |
| Inflammatory Response | Absent | Present |
| Energy Requirement | Energy-dependent, ATP-requiring | Energy-independent |
| Genomic Control | Genetically regulated | Not genetically controlled |
The distinction between apoptosis and necrosis is fundamental in cell death research. While apoptosis is a tightly regulated, energy-dependent process [3], necrosis is an uncontrolled, passive process typically resulting from acute cellular injury [2]. Morphologically, necrosis is characterized by cell swelling, formation of cytosolic vacuoles, distended endoplasmic reticulum, swollen or ruptured mitochondria, and eventual cell membrane disruption [3]. This membrane rupture results in the release of cytoplasmic contents into the surrounding environment, triggering inflammatory responses [3].
Molecular Mechanisms of Apoptosis
The morphological changes observed during apoptosis result from the precise activation and execution of molecular pathways. The two primary initiation routes are the extrinsic (death receptor) pathway and the intrinsic (mitochondrial) pathway, both culminating in the activation of caspases that execute the cell death program.
The Extrinsic Pathway
The extrinsic pathway is initiated by the binding of extracellular death ligands to their corresponding transmembrane death receptors, which belong to the tumor necrosis factor (TNF) receptor superfamily [3]. Notable death ligands and receptors include FasL/FasR, TNF-α/TNFR1, Apo3L/DR3, and Apo2L/DR4/DR5 [3].
Upon ligand binding, the receptors undergo trimerization and recruit intracellular adapter proteins such as FADD (Fas-associated protein with death domain) through protein-protein interactions mediated by death domains [3]. This complex, known as the death-inducing signaling complex (DISC), recruits and activates initiator caspases (primarily caspase-8 and caspase-10) through proximity-induced autocatalytic cleavage [2] [3]. Active caspase-8 then activates executioner caspases (caspase-3, -6, and -7), which systematically cleave cellular substrates to bring about the morphological changes characteristic of apoptosis [2].
Diagram 1: Extrinsic apoptosis pathway activation
The Intrinsic Pathway
The intrinsic pathway, also known as the mitochondrial pathway, is triggered by intracellular stress signals such as DNA damage, oxidative stress, growth factor withdrawal, or endoplasmic reticulum stress [2]. These signals cause the Bcl-2 protein family to regulate mitochondrial outer membrane permeabilization (MOMP) [1].
Pro-apoptotic Bcl-2 family proteins (such as Bax and Bak) oligomerize and form pores in the mitochondrial outer membrane, while anti-apoptotic members (including Bcl-2 and Bcl-xL) inhibit this process [2]. The permeabilization of the mitochondrial membrane leads to the release of several pro-apoptotic proteins from the intermembrane space into the cytosol, including cytochrome c and SMAC (second mitochondria-derived activator of caspases) [2].
Cytochrome c binds to Apaf-1 (apoptotic protease activating factor 1) and ATP to form the apoptosome, which recruits and activates procaspase-9 [2]. Active caspase-9 then activates the executioner caspases (primarily caspase-3). Simultaneously, SMAC proteins neutralize inhibitor of apoptosis proteins (IAPs), thereby relieving their suppression of caspase activity [2].
Diagram 2: Intrinsic apoptosis pathway mechanism
Caspase Activation and Execution
Both the intrinsic and extrinsic pathways converge on the activation of executioner caspases (primarily caspase-3, -6, and -7), which orchestrate the systematic dismantling of cellular structures [2]. Caspases are cysteine proteases that cleave their substrates after aspartic acid residues [3]. Their activation initiates a proteolytic cascade that amplifies the apoptotic signal and ensures rapid, irreversible commitment to cell death.
Executioner caspases target hundreds of cellular proteins, including:
- Nuclear proteins: Lamin proteins (nuclear envelope disintegration), ICAD/DFF45 (activation of DNases causing DNA fragmentation)
- Cytoskeletal proteins: Actin, fodrin, gelsolin (membrane blebbing and apoptotic body formation)
- DNA repair enzymes: Poly (ADP-ribose) polymerase (PARP)
- Cell cycle regulators
- Cell adhesion molecules
This targeted proteolysis results in the characteristic morphological changes of apoptosis while maintaining membrane integrity to prevent inflammatory responses [1] [3].
Experimental Methods for Apoptosis Detection
Accurate detection and quantification of apoptosis are essential for research and therapeutic development. Multiple methodologies exist that target different aspects of the apoptotic process, from morphological assessment to biochemical and molecular analyses.
Morphological Assessment Techniques
Light and Electron Microscopy: The initial identification of apoptotic cells often relies on recognizing characteristic morphological changes using various microscopic techniques [3]. Light microscopy can reveal cell shrinkage, chromatin condensation, and apoptotic body formation in stained tissue sections [3]. Electron microscopy provides higher resolution details, including organelle integrity, chromatin margination, and membrane blebbing [3].
Time-Lapse Microscopy: Live-cell imaging allows for the dynamic observation of apoptotic progression in real-time, including membrane blebbing, cell shrinkage, and apoptotic body formation [3].
Biochemical and Molecular Detection Methods
Table 3: Key Experimental Methods for Apoptosis Detection
| Method | Principle | Application | Advantages | Limitations |
|---|---|---|---|---|
| TUNEL Assay | Detects DNA fragmentation by labeling 3'-OH ends | In situ detection of apoptotic cells in tissue sections | High sensitivity, works in fixed tissues | Can detect non-apoptotic DNA damage [1] |
| DNA Laddering | Agarose gel electrophoresis of fragmented DNA | Detection of internucleosomal DNA cleavage | Classic apoptosis confirmation | Requires many cells, not quantitative [1] |
| Caspase Activity Assays | Fluorogenic or colorimetric substrate cleavage | Measurement of caspase activation | Quantitative, specific to apoptosis | Does not indicate late-stage apoptosis |
| Annexin V Staining | Binds to phosphatidylserine externalized on membrane | Detection of early apoptotic stages | Distinguishes early vs late apoptosis | Requires live cells, confounded by necrosis |
| Mitochondrial Membrane Potential | Fluorescent dyes (JC-1, TMRM) | Assessment of mitochondrial integrity in intrinsic pathway | Early indicator of intrinsic apoptosis | Not specific to apoptosis alone |
| Western Blotting | Detection of cleavage products (PARP, caspases) | Confirmation of apoptotic protein activation | Specific, provides molecular evidence | Semi-quantitative, requires protein extraction |
The TUNEL (TdT-mediated dUTP nick-end labeling) assay is a widely used method for detecting apoptotic cells in tissue samples by labeling the 3'-OH ends of fragmented DNA [1]. TUNEL-positive cardiomyocytes show morphological features of apoptosis and the typical ladder pattern in DNA electrophoresis [1]. However, careful standardization of the staining protocol is essential, as the assay can detect non-apoptotic DNA damage under suboptimal conditions [1].
Caspase activity assays utilize fluorogenic or colorimetric substrates that emit signals upon cleavage by active caspases, providing quantitative data on apoptotic progression [1]. These assays can be adapted for high-throughput screening of potential therapeutic compounds that modulate apoptosis.
Annexin V staining capitalizes on the externalization of phosphatidylserine during early apoptosis, which serves as an "eat-me" signal for phagocytes [3]. When combined with viability dyes like propidium iodide, this method can distinguish between early apoptotic (Annexin V+/PI-), late apoptotic (Annexin V+/PI+), and necrotic (Annexin V-/PI+) cells [3].
Research Reagent Solutions
The investigation of apoptotic mechanisms relies on a comprehensive toolkit of research reagents that enable the specific detection, modulation, and analysis of cell death pathways.
Table 4: Essential Research Reagents for Apoptosis Studies
| Reagent Category | Specific Examples | Research Application | Mechanism of Action |
|---|---|---|---|
| Caspase Inhibitors | Z-VAD-FMK (pan-caspase inhibitor) | Inhibition of apoptotic execution | Irreversible binding to active site of caspases [4] |
| Death Receptor Ligands | Recombinant TNF-α, FasL, TRAIL | Extrinsic pathway activation | Binding to death receptors to initiate DISC formation [3] |
| Bcl-2 Family Modulators | ABT-737 (Bcl-2 inhibitor), AT-101 (Bcl-2/Bcl-xL inhibitor) | Modulating intrinsic pathway | Disrupting anti-apoptotic protein function [2] |
| IAP Antagonists | Smac mimetics (BV6) | Sensitizing cells to apoptosis | Neutralizing IAP proteins to promote caspase activation [5] [4] |
| Kinase Inhibitors | Cabozantinib (Met inhibitor), Necrostatin-1 (RIPK1 inhibitor) | Pathway-specific modulation | Targeting regulatory kinases in apoptotic signaling [4] |
| Mitochondrial Dyes | JC-1, TMRM, MitoTracker | Assessing mitochondrial health | Indicators of mitochondrial membrane potential [3] |
| Apoptosis Inducers | Staurosporine, Actinomycin D, Etoposide | Experimental induction of apoptosis | DNA damage or kinase inhibition [2] |
| Detection Antibodies | Anti-cleaved caspase-3, anti-PARP, anti-Bax | Immunodetection of apoptotic markers | Recognizing specific epitopes on apoptotic proteins [1] |
These research reagents enable precise investigation of apoptotic mechanisms and potential therapeutic interventions. For example, Smac mimetics are cytotoxic agents specifically designed to maximize tumor cell killing mediated via endogenous tumor necrosis factor (TNF) by targeting IAP proteins for degradation [4]. Similarly, caspase inhibitors like ZVAD allow researchers to distinguish between apoptotic and non-apoptotic cell death mechanisms and have been instrumental in identifying hybrid cell death processes [4].
Advanced Research Applications
Crosstalk Between Cell Death Mechanisms
Emerging research reveals extensive crosstalk between different cell death mechanisms, including apoptosis, autophagy, ferroptosis, necroptosis, mitophagy, and pyroptosis [6]. This crosstalk enables cells to integrate diverse stress signals and determine the most appropriate death modality based on cellular context, energy status, and environmental factors.
Key nodes in cell death crosstalk include:
- Caspase-8: Functions as a molecular switch between apoptosis and necroptosis
- Bcl-2 family proteins: Regulate both apoptosis and autophagy
- Mitochondrial integrity: Central to apoptosis, necroptosis, and pyroptosis
- Reactive oxygen species: Common inducers of multiple cell death pathways
Understanding these interconnections provides novel therapeutic opportunities, particularly for overcoming treatment resistance in cancer, where tumor cells often develop defects in apoptotic pathways [6] [4].
Computational Modeling of Apoptosis
Boolean or logical modeling has emerged as a promising approach to capture the qualitative behavior of complex apoptotic networks [5]. These models represent the apoptotic signaling network as a series of logical operations (ON/OFF states) that respond to various inputs such as Fas ligand, TNF-α, UV-B irradiation, and other stimuli [5].
Advanced Boolean models of apoptosis incorporate 86 nodes and 125 interactions, utilizing timescales and multi-value node logic to reproduce dynamic features such as threshold behavior, feedback loops, and reaction delays [5]. These computational approaches help identify critical regulatory hubs in the apoptotic network and predict cellular responses to combinatorial treatments, facilitating the development of selective control strategies for pathological conditions [7].
Therapeutic Targeting of Apoptosis Pathways
The precise regulation of apoptosis has significant therapeutic implications across multiple disease areas. In cancer, where apoptosis is often repressed, strategies focus on restoring or enhancing apoptotic sensitivity through:
- BH3 mimetics that inhibit anti-apoptotic Bcl-2 family proteins
- Smac mimetics that neutralize IAP-mediated caspase inhibition
- Death receptor agonists that directly activate extrinsic apoptosis
- Combination therapies that simultaneously target multiple regulatory nodes
Recent research demonstrates that targeting the Met-RIPK1 signaling axis with Cabozantinib can sensitize colorectal cancer cells to Smac mimetic-induced apoptosis and necroptosis, providing a promising approach to overcome therapy resistance [4]. Similarly, modulating between apoptosis and necroptosis represents a strategic approach to maximize tumor cell killing and foster anti-tumor immunity [4].
Apoptosis represents a critically important programmed cell death process characterized by distinctive morphological features that result from the precise execution of molecular pathways. The systematic investigation of apoptotic mechanisms—from initial morphological observations to current understanding of complex signaling networks—has provided fundamental insights into cellular homeostasis and disease pathogenesis. Advanced research methodologies, including sophisticated detection assays, targeted research reagents, and computational modeling approaches, continue to enhance our understanding of apoptotic regulation and its interconnections with other cell death modalities. This comprehensive knowledge base provides the foundation for developing novel therapeutic strategies that selectively modulate cell death pathways in cancer, neurodegenerative disorders, and other pathological conditions, ultimately advancing the frontier of precision medicine.
Phase I (Early Apoptosis) represents the initial and commitment stage of programmed cell death, characterized by a defined set of morphological alterations that precede the complete dismantling of the cell. These early changes—cell shrinkage, cytoplasmic condensation, and loss of microvilli—serve as the first visible indicators that the apoptotic cascade has been irreversibly activated [8] [9]. This phase is distinct from accidental cell death (necrosis) and is tightly regulated by molecular machinery that transforms the cell's structure with remarkable precision [10]. The events of Phase I are not passive degenerative processes but are actively executed by proteases and other enzymes, setting the stage for subsequent phases involving nuclear fragmentation and the formation of apoptotic bodies [8].
Understanding these initial morphological hallmarks is crucial for researchers and drug development professionals. They provide a foundation for identifying apoptotic cells in experimental and clinical samples, from tissue sections to cell cultures, and are essential for validating the efficacy of therapies designed to modulate cell death, such as in cancer treatment [11]. This guide provides a detailed technical examination of the defining features, underlying mechanisms, and detection methodologies for Phase I apoptosis.
Morphological Hallmarks of Phase I Apoptosis
The transition of a cell into Phase I apoptosis involves a coordinated series of structural changes. The following table summarizes the core morphological features and their functional consequences.
Table 1: Core Morphological Features of Phase I Apoptosis
| Morphological Feature | Description | Functional Consequence |
|---|---|---|
| Cell Shrinkage | Reduction in cell volume and disruption of the cytoskeleton, leading to a smaller, more condensed cellular profile [8]. | Represents the initial break with normal cellular homeostasis and is a key feature distinguishing apoptosis from necrotic cell swelling [10]. |
| Cytoplasmic Condensation | Increased density of the cytoplasmic matrix and organelles, with the cell becoming deeply eosinophilic in stained preparations [8]. | Results from caspase-mediated cleavage of structural proteins and dehydration, concentrating the cellular contents. |
| Loss of Microvilli and Cell-Cell Contact | Breakdown of specialized surface structures, including microvilli, and detachment from neighboring cells and the extracellular matrix [8] [9]. | Facilitates the isolation of the dying cell from its healthy neighbors, a prelude to its eventual removal. |
The following diagram illustrates the temporal relationship and key signaling initiators of these morphological events during Phase I apoptosis:
Molecular Mechanisms and Key Players
The dramatic structural changes observed in Phase I are the direct result of the activation of caspases, a family of cysteine-aspartic proteases that act as the central executioners of apoptosis [8] [9]. Initiator caspases (e.g., caspase-8, -9) are activated upstream by either the extrinsic (death receptor) or intrinsic (mitochondrial) pathways. These, in turn, activate the executioner caspases-3, -6, and -7 [8] [10]. Caspase-3 is particularly crucial and targets several key structural proteins to initiate Phase I morphology:
- Cytoskeletal Disassembly: Caspase-3 cleaves proteins such as gelsolin, which then severs actin filaments, and ROCK1 kinase, which induces membrane blebbing by causing actomyosin contraction [9]. The breakdown of the cytoskeletal framework is the primary driver of cell shrinkage and the loss of structural integrity.
- Loss of Cell Adhesion: The cleavage of focal adhesion kinases (FAKs) and other adhesion complex proteins contributes to the loss of cell-cell and cell-matrix contacts, explaining the observed rounding and detachment of apoptotic cells [8].
Concurrently, one of the earliest biochemical events, which often precedes overt morphological changes, is the translocation of phosphatidylserine (PS) from the inner to the outer leaflet of the plasma membrane [12]. This "eat-me" signal is recognized by phagocytic cells and is facilitated by the caspase-mediated inactivation of the flippase ATP11A and activation of the scramblase Xkr8 [9].
Table 2: Key Molecular Players in Phase I Apoptosis Morphology
| Molecule | Role/Function in Phase I | Effect on Morphology |
|---|---|---|
| Caspase-3 | Primary executioner caspase; cleaves numerous cellular substrates [9]. | Orchestrates multiple morphological changes including shrinkage, condensation, and membrane blebbing. |
| Gelsolin | Actin-binding protein; cleaved and activated by caspases [8]. | Severs actin filaments, leading to the dissolution of the cytoskeleton and cell shrinkage. |
| ROCK1 | Kinase regulating actomyosin contraction; cleaved and activated by caspases [9]. | Induces forceful contraction of the cell cortex, resulting in membrane blebbing. |
| Xkr8 / ATP11A | Plasma membrane phospholipid scramblase and flippase, respectively [9]. | Regulates phosphatidylserine externalization, a key "eat-me" signal for phagocytes. |
Experimental Detection and Analysis Protocols
Detecting Phase I apoptosis requires assays that capture the initial structural and membrane changes. A multi-modal approach is recommended for robust confirmation [8].
Light and Electron Microscopy
Protocol: Morphological Assessment via Microscopy
- Sample Preparation: Culture cells or collect tissue samples. Induce apoptosis using a relevant stimulus (e.g., staurosporine, chemotherapeutic agent, UV irradiation). Fix cells with 4% paraformaldehyde [10].
- Staining:
- Analysis: Examine slides under a light microscope. Apoptotic cells are identified by their reduced size, condensed cytoplasm, and nuclear changes (chromatin condensation) [8]. For higher resolution, transmission electron microscopy (TEM) can be used to confirm organelle condensation and loss of microvilli [9].
Annexin V Staining for Phosphatidylserine Exposure
Protocol: Flow Cytometry for Early Apoptosis
- Cell Harvesting: Gently harvest adherent cells using a non-enzymatic dissociation buffer to preserve membrane integrity [12].
- Staining:
- Incubation and Analysis: Incubate cells for 15-20 minutes in the dark. Analyze immediately using a flow cytometer.
- Early Apoptotic Cells: Annexin V positive / PI negative.
- This assay is a cornerstone for quantifying cells in the early stages of apoptosis [12].
Analysis of Caspase Activation
Protocol: Immunoblotting for Caspase-3 Cleavage
- Protein Extraction: Lyse control and treated cells in RIPA buffer containing protease inhibitors.
- Electrophoresis and Transfer: Separate proteins via SDS-PAGE (12-15% gel) and transfer to a nitrocellulose or PVDF membrane.
- Immunodetection:
- Block membrane with 5% non-fat milk.
- Probe with a primary antibody against cleaved caspase-3 (preferable) or total caspase-3.
- Incubate with an HRP-conjugated secondary antibody and detect using chemiluminescence [10].
- Expected Result: Appearance of a lower molecular weight band (~17/19 kDa) corresponding to the active subunits of caspase-3 in apoptotic samples, confirming the activation of the executioner phase [8].
The following workflow diagram integrates these key methodologies into a coherent experimental strategy:
The Scientist's Toolkit: Essential Research Reagents
A range of well-characterized reagents is critical for the experimental investigation of Phase I apoptosis. The following table details essential tools and their applications.
Table 3: Key Research Reagents for Studying Phase I Apoptosis
| Reagent/Category | Specific Examples | Function and Application |
|---|---|---|
| Viability & Membrane Assays | Annexin V (FITC, PE conjugates); Propidium Iodide (PI); 7-AAD | Detects phosphatidylserine exposure and loss of membrane integrity to distinguish early and late apoptotic stages via flow cytometry [8] [12]. |
| Caspase Activity Assays | Fluorogenic substrates (e.g., DEVD-AFC for caspase-3); Caspase inhibitors (e.g., Z-VAD-FMK) | Measures enzymatic activity of caspases for proof of apoptotic mechanism; inhibitors confirm caspase-dependency of observed death [8]. |
| Antibodies for Immunoassay | Anti-cleaved caspase-3; Anti-cytochrome c; Anti-Bcl-2 family proteins | Used in Western blot (WB) and Immunohistochemistry (IHC) to detect activation of key apoptotic proteins and regulators [12] [10]. |
| DNA-Binding Dyes | Hoechst 33342; DAPI; Acridine Orange | Stain condensed chromatin in apoptotic nuclei for visualization by fluorescence microscopy [8]. |
| Inducers/Inhibitors | Staurosporine; TRAIL; ABT-263 (Navitoclax); TNF-α | Pharmacological tools to reliably induce apoptosis (intrinsic/extrinsic pathways) or inhibit specific anti-apoptotic proteins (e.g., Bcl-2) for experimental control [13] [11]. |
The events of Phase I apoptosis—cell shrinkage, cytoplasmic condensation, and loss of microvilli—are the definitive morphological signature of a cell undergoing programmed demolition. These changes are not passive but are actively driven by the precise cleavage of structural proteins by activated caspases. Mastery of the assays to detect these changes, from Annexin V staining to caspase immunoblotting, is fundamental for research in cell biology, toxicology, and drug development. As therapeutic strategies increasingly aim to modulate apoptosis, particularly in oncology [11], a rigorous understanding of this initial phase provides the critical framework for analyzing therapeutic efficacy and understanding resistance mechanisms.
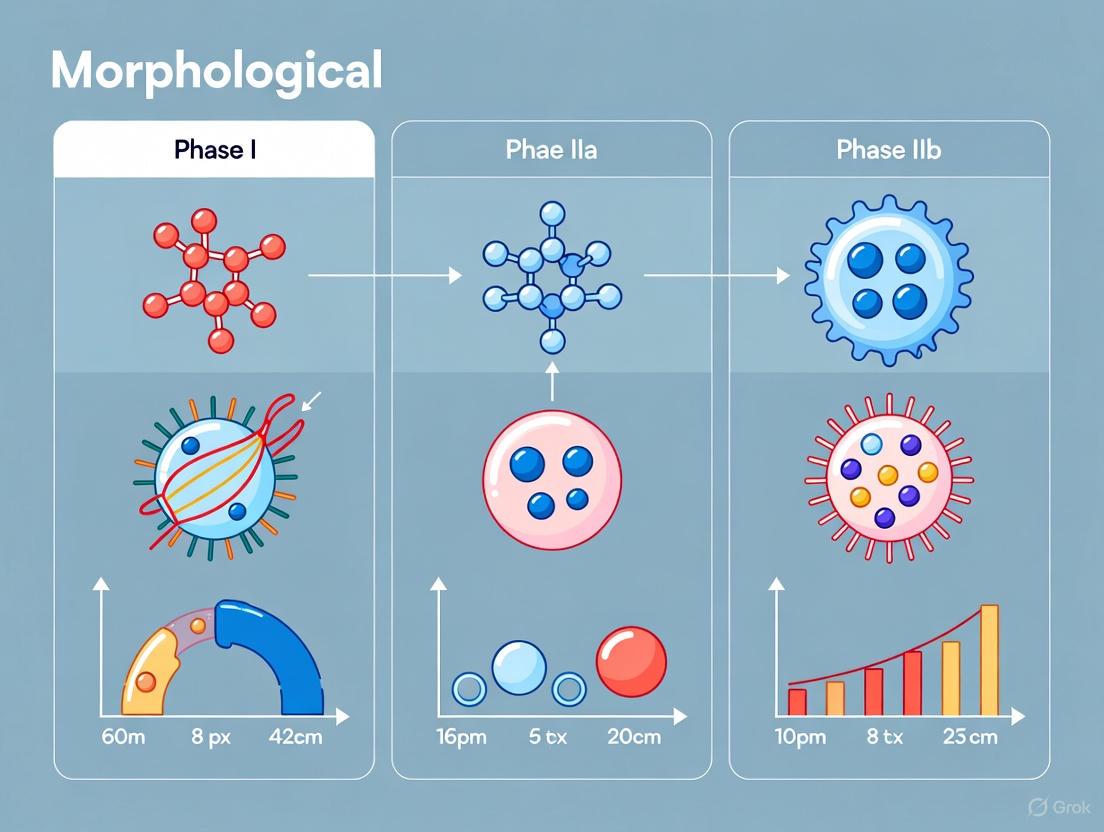
Within the broader morphological framework of apoptosis, cell death progresses through three distinct nuclear phases: Phase I (chromatin condensation and cell shrinkage), Phase IIa (nuclear collapse characterized by pyknosis and chromatin margination), and Phase IIb (nuclear fragmentation into apoptotic bodies) [14]. This whitepaper provides an in-depth technical examination of Phase IIa apoptosis, a critical middle stage marked by definitive nuclear collapse. During this stage, the cell commits to the point of no return in the death pathway [14]. We will delineate the characteristic morphological features of Phase IIa, detail the molecular mechanisms driving these changes, and present robust experimental protocols for its detection and quantification. A thorough understanding of this phase is paramount for basic cell biology research and for the development of therapeutics designed to induce or inhibit cell death in diseases such as cancer [15].
Morphological and Nuclear Characteristics of Phase IIa Apoptosis
Phase IIa represents the stage of nuclear collapse and disassembly, serving as a bridge between the initial condensation of Phase I and the final packaging of cellular contents into apoptotic bodies in Phase IIb [14]. The defining morphological characteristics of this stage are profound and observable at the ultrastructural level.
The most prominent feature is chromatin condensation, where the nuclear chromatin becomes densely packed [6]. This is closely followed by pyknosis, the irreversible condensation of the nuclear chromatin resulting in a reduction of nuclear size and increased basophilia, and nuclear margination, a process where the condensed chromatin aggregates along the inner periphery of the nuclear membrane [14] [15]. Concurrently, the cell itself continues to shrink and undergoes a process of budding, and the cytoskeleton begins to degrade [14]. It is critical to note that the integrity of the plasma membrane is maintained throughout this phase, preventing the release of intracellular contents and an inflammatory response, which distinguishes apoptosis from necrotic cell death [14] [6].
Table 1: Key Morphological Characteristics of Apoptosis Phase IIa
| Feature | Description | Technical Observation Method |
|---|---|---|
| Chromatin Condensation | Chromatin becomes highly compacted and densely stained. | Electron microscopy; Fluorescence microscopy (DAPI/Hoechst) [14] |
| Pyknosis | Irreversible condensation of nuclear chromatin, leading to a small, dense nucleus. | Light microscopy (HE staining); Fluorescence microscopy [15] |
| Nuclear Margination | Condensed chromatin aggregates on the inner nuclear membrane. | Electron microscopy; Fluorescence microscopy [14] |
| Nuclear Shrinkage | Overall reduction in nuclear volume. | Computerized morphometric analysis of stained nuclei [15] |
| Intact Plasma Membrane | Cellular membrane remains intact, preventing inflammatory response. | Exclusion dyes (e.g., Propidium Iodide) in live cells [14] |
Molecular Mechanisms and Signaling Pathways
The dramatic nuclear morphology of Phase IIa is executed by a tightly regulated molecular cascade, primarily driven by the activation of caspases and specific endonucleases.
Caspase Activation and Substrate Cleavage
The apoptotic process, including the transition to Phase IIa, is dependent on the activation of a family of cysteine proteases known as caspases [16]. These exist as inactive zymogens in healthy cells and are cleaved and activated in a cascade. Both the intrinsic (mitochondrial) and extrinsic (death receptor) pathways converge on the activation of executioner caspases, such as caspase-3 and caspase-7 [17] [6]. Once activated, these caspases cleave a wide array of intracellular substrates, including structural proteins of the nucleus and cytoskeleton, which facilitates the morphological changes characteristic of apoptosis [17].
The Role of DFF40/CAD Endonuclease in Chromatin Collapse
A key substrate of caspase-3 is DFF45/ICAD (Inhibitor of Caspase-Activated DNase), the chaperone and inhibitor of the endonuclease DFF40 (also known as CAD) [16]. Cleavage of DFF45/ICAD leads to the release and activation of DFF40/CAD [16]. While this endonuclease is famously known for hydrolyzing DNA into oligonucleosomal-sized fragments (the DNA ladder), research indicates that its role in Phase IIa nuclear collapse is distinct.
Studies using cell models that undergo caspase-dependent apoptosis without DNA laddering have shown that DFF40/CAD is still essential for the chromatin compaction and nuclear disassembly of Phase IIa [16]. The mechanism involves DFF40/CAD generating single-strand DNA nicks/breaks (SSBs) with 3'-OH ends, rather than the double-strand breaks responsible for the DNA ladder [16]. This specific type of DNA damage is sufficient to prompt the highest order of chromatin compaction observed in Stage II apoptotic nuclei. Therefore, Phase IIa chromatin collapse relies on DFF40/CAD-mediated DNA damage, with the nature of the DNA break being a critical factor.
The following diagram illustrates the key molecular events leading to Phase IIa morphology:
Experimental Protocols for Detection and Analysis
Accurate identification and quantification of Phase IIa apoptosis require a combination of morphological, biochemical, and cytometric techniques. Below are detailed protocols for key methodologies.
Fluorescence Microscopy and Nuclear Morphometry
This protocol allows for the quantitative assessment of apoptotic nuclear changes, including pyknosis and shrinkage [15].
Procedure:
- Cell Culture and Induction: Seed cells (e.g., LNCaP or MDA-MB-231) onto culture dishes or multi-well plates and allow them to adhere. Treat with an apoptosis-inducing agent (e.g., 3.0 μM Cycloheximide) for a predetermined time (e.g., 24 hours) [15].
- Fixation and Permeabilization: Wash cells twice with Phosphate Buffered Saline (PBS). Permeabilize cells by incubating with 0.2% Triton X-100 in PBS for 10-15 minutes at room temperature [15].
- Nuclear Staining: Wash cells with PBS. Incubate with a nuclear stain, such as 4',6-diamidino-2-phenylindole (DAPI) at a concentration of 1.0 μg/ml, for 10-20 minutes protected from light [15].
- Image Acquisition: Using a fluorescence microscope (e.g., Keyence BZ-9000), acquire images from multiple random fields using a DAPI filter set with a 20x or higher magnification objective [15].
- Morphometric Analysis: Use image analysis software (e.g., BZ II Analyzer) to quantify parameters for each nucleus. Key parameters include:
- Area: The total area of the nucleus (μm²). Apoptotic nuclei show a significant decrease.
- Perimeter: The outer boundary length of the nucleus (μm).
- Major and Minor Axis: The longest and shortest diameter of the nucleus (μm).
- Brightness: The mean fluorescence intensity. Apoptotic nuclei often show increased brightness due to chromatin condensation [15].
Table 2: Key Research Reagents for Phase IIa Apoptosis Analysis
| Reagent / Tool | Function / Application | Experimental Context |
|---|---|---|
| DAPI / Hoechst 33258 | Fluorescent DNA dyes that bind preferentially to A-T regions, staining the nucleus. | Visualization of nuclear morphology (condensation, pyknosis) via fluorescence microscopy [15]. |
| Cycloheximide (CHX) | Inhibitor of protein synthesis; a potent activator of apoptotic pathways. | Used as a positive control inducer of apoptosis in model cell lines [15]. |
| Caspase Inhibitor (e.g., Q-VD-OPh) | Pan-caspase inhibitor that prevents the activation of executioner caspases. | Tool to confirm the caspase-dependence of the observed nuclear morphology [16]. |
| Anti-DFF40/CAD Antibody | Specific antibody for immunoblotting or immunofluorescence. | Used to detect the expression and cleavage/activation status of the DFF40/CAD endonuclease [16]. |
| TUNEL Assay Kit | Labels 3'-OH ends of DNA fragments with fluorescent tags. | Detects the DNA strand breaks generated during apoptosis, including those in Phase IIa [16] [15]. |
In Situ Detection of DNA Strand Breaks (TUNEL Assay)
The TUNEL (TdT dUTP Nick-End Labeling) assay is a key method for detecting the 3'-OH DNA ends generated by DFF40/CAD and other nucleases during apoptosis [16].
Procedure:
- Cell Preparation and Fixation: Culture and induce apoptosis in cells grown on glass coverslips. Fix cells with 4% paraformaldehyde for 15-30 minutes at room temperature.
- Permeabilization: Permeabilize the fixed cells with a mild detergent (e.g., 0.1% Triton X-100 in PBS) for 5-10 minutes on ice to allow enzyme access to the nucleus.
- Labeling Reaction: Prepare the TUNEL reaction mixture containing Terminal Deoxynucleotidyl Transferase (TdT) and fluorescently-labeled dUTP (e.g., FITC-dUTP). Incubate the coverslips with the reaction mixture in a humidified chamber for 60 minutes at 37°C in the dark.
- Counterstaining and Mounting: Wash the coverslips thoroughly to remove unincorporated nucleotides. Counterstain the nuclei with DAPI or Hoechst to visualize all cells.
- Analysis: Mount the coverslips and analyze by fluorescence microscopy. Cells with positive TUNEL staining (green fluorescence) indicate the presence of DNA strand breaks, a hallmark of mid-to-late stage apoptosis. It is crucial to include appropriate controls (e.g., DNase-treated cells as a positive control, and omission of TdT enzyme as a negative control) [14] [16].
Discriminating Apoptosis from Necrosis in Real-Time
Advanced live-cell imaging techniques can dynamically distinguish Phase IIa apoptosis from necrotic cell death, which is vital for understanding drug mechanisms.
Procedure Utilizing FRET-Based Caspase Sensor:
- Stable Cell Line Generation: Engineer a cell line (e.g., U251 neuroblastoma) to stably express two constructs: a soluble FRET-based caspase sensor (e.g., ECFP-DEVD-EYFP) and a non-soluble organelle-targeted fluorescent protein (e.g., Mito-DsRed) [18].
- Real-Time Imaging: Treat the cells with the compound of interest and perform time-lapse imaging using a fluorescence microscope equipped with environmental control (37°C, 5% CO₂).
- Data Interpretation:
- Viable Cells: Exhibit intact FRET (yellow emission) and mitochondrial red fluorescence.
- Apoptotic Cells (Phase IIa): Show a loss of FRET (increase in blue donor emission) due to caspase-mediated cleavage of the DEVD linker, while retaining mitochondrial DsRed fluorescence.
- Necrotic Cells: Lose the soluble FRET probe completely (no ECFP or EYFP signal) due to membrane rupture, while initially retaining the mitochondrial DsRed signal [18].
The experimental workflow for a multi-method approach to characterizing Phase IIa apoptosis is summarized below:
Phase IIa apoptosis, characterized by chromatin condensation, pyknosis, and nuclear margination, represents a decisive commitment to cell death driven by caspase-3 and the DFF40/CAD-mediated generation of single-strand DNA breaks. A comprehensive approach, utilizing the quantitative and qualitative methods detailed in this guide, is essential for researchers to accurately identify and study this critical phase. As our understanding of the molecular crosstalk between different cell death pathways deepens [6], the precise characterization of apoptotic stages will become increasingly important for developing more effective and targeted therapeutic strategies, particularly in oncology and neurodegenerative diseases.
Phase IIb, or late apoptosis, represents the terminal executive phase of the programmed cell death process, characterized by systematic cellular disintegration. This phase follows the initial signaling events and early morphological changes, culminating in the hallmark features of nuclear fragmentation and apoptotic body formation [17] [19]. These structural changes represent the irreversible commitment to cell death and facilitate the safe packaging and removal of cellular debris without eliciting an inflammatory response, distinguishing apoptosis from necrotic cell death [17] [13].
The biological significance of these late-stage events lies in their role in maintaining tissue homeostasis. By efficiently disposing of unwanted cells through phagocytosis by neighboring cells or professional phagocytes, apoptosis prevents the release of intracellular contents that could trigger inflammation or autoimmune reactions [6] [13]. This silent elimination is particularly crucial during developmental processes, tissue remodeling, and the elimination of damaged or potentially harmful cells [19].
Morphological Hallmarks of Phase IIb
Nuclear Fragmentation
Nuclear fragmentation, also known as karyorrhexis, involves the systematic breakdown of the nucleus into discrete, membrane-bound fragments. This process begins with chromatin condensation, where nuclear chromatin aggregates into dense, marginalized masses against the nuclear envelope [17] [19]. The nuclear envelope then invaginates and fragments, followed by the separation of the condensed chromatin into multiple discrete nuclear bodies [17].
This nuclear disintegration is mediated by the activation of specific endonucleases, particularly caspase-activated DNase (CAD), which cleaves DNA at internucleosomal regions, producing characteristic DNA fragments in multiples of approximately 180-200 base pairs [19]. This cleavage pattern results in the distinctive DNA laddering pattern observed in gel electrophoresis, which serves as a biochemical hallmark of apoptosis [19].
Apoptotic Body Formation
Following nuclear disintegration, the cell undergoes a coordinated process of segmentation into apoptotic bodies. The cell membrane undergoes pronounced blebbing, forming protrusions that eventually separate from the main cell body [20] [19]. These membrane-bound vesicles typically range from 0.5 to 2.0 micrometers in diameter and contain various cellular components, including intact organelles, nuclear fragments, and cytoplasmic elements [17].
Critically, during this process, phosphatidylserine—a phospholipid normally restricted to the inner leaflet of the plasma membrane—translocates to the external surface of the apoptotic bodies [17]. This surface alteration serves as an "eat me" signal for phagocytic cells, facilitating the recognition and clearance of the apoptotic debris [17] [19]. The entire process occurs without compromising plasma membrane integrity, thus preventing the release of pro-inflammatory intracellular components [13].
Table 1: Key Morphological Features of Phase IIb Apoptosis
| Morphological Feature | Description | Molecular Mediators | Functional Significance |
|---|---|---|---|
| Chromatin Condensation | Aggregation of nuclear chromatin into dense, marginalized masses | Histone modification, caspase activation | Inactivates genetic material, initiates nuclear breakdown |
| Nuclear Fragmentation | Disintegration of nucleus into multiple discrete fragments | Caspase-activated DNase (CAD), Lamin cleavage | Packages nuclear material for disposal |
| DNA Fragmentation | Cleavage at internucleosomal regions | Endonucleases | Produces characteristic DNA laddering pattern |
| Membrane Blebbing | Protrusion and bulging of plasma membrane | ROCK1-mediated actin cytoskeleton reorganization | Facilitates cell segmentation |
| Apoptotic Body Formation | Formation of membrane-bound vesicles containing cellular components | Cytoskeletal breakdown, membrane remodeling | Packages cellular contents for phagocytosis |
| Phosphatidylserine Externalization | Translocation to outer membrane leaflet | Scramblase activation, floppase inhibition | Promotes recognition by phagocytic cells |
Quantitative Analysis of Morphological Changes
Advanced imaging technologies have enabled precise quantification of the morphological alterations characterizing late apoptosis. Studies utilizing high-resolution techniques like Full-Field Optical Coherence Tomography (FF-OCT) have documented consistent dimensional changes during this phase [20].
Cells undergoing late apoptosis demonstrate a significant reduction in cell volume—typically decreasing to 40-60% of their original size—as the cytoplasm condenses and organelles are packaged into apoptotic bodies [20]. The nuclear-to-cytoplasmic ratio also decreases dramatically as the nucleus fragments and disperses. Time-lapse imaging reveals that the process from initial nuclear condensation to complete apoptotic body formation typically occurs within 30-180 minutes, depending on cell type and apoptotic stimulus [20].
Table 2: Quantitative Parameters of Late Apoptosis Morphology
| Parameter | Measurement Method | Typical Values in Late Apoptosis | Technical Notes |
|---|---|---|---|
| Cell Volume Reduction | FF-OCT 3D topography | 40-60% of original volume | Measured via surface reconstruction |
| Apoptotic Body Size | Electron microscopy, FF-OCT | 0.5-2.0 μm diameter | Membrane-bound vesicles |
| Nuclear Condensation | Chromatin staining intensity | 2-3 fold increase in density | DAPI/Hoechst fluorescence |
| DNA Fragmentation | Gel electrophoresis | 180-200 bp multiples | "DNA laddering" pattern |
| Time Course | Live-cell imaging | 30-180 minutes | Cell type and stimulus dependent |
| Phosphatidylserine Exposure | Annexin V binding | >80% of cells | Detected before membrane permeability |
Molecular Mechanisms and Signaling Pathways
The morphological changes of Phase IIb apoptosis are executed through the coordinated activation of specific molecular pathways. The caspase cascade serves as the central executioner, with initiator caspases (caspase-8, -9) activating effector caspases (caspase-3, -6, -7) that directly cleave cellular structural proteins [6] [17] [19].
Nuclear Disassembly Mechanisms
Nuclear fragmentation is mediated through the caspase-mediated cleavage of key nuclear structural proteins. Lamin proteins, which form the nuclear lamina scaffolding, are cleaved by caspase-6, leading to the collapse of the nuclear envelope integrity [17]. Simultaneously, activation of caspase-activated DNase (CAD) through cleavage of its inhibitor (ICAD) by caspase-3 results in DNA fragmentation at internucleosomal sites [19]. Additional caspase targets include proteins involved in DNA repair (such as PARP) and nuclear transport, ensuring the systematic dismantling of nuclear function and structure [6].
Cytoskeletal Reorganization and Membrane Blebbing
The dramatic changes in cell shape and the formation of apoptotic bodies are driven by caspase-mediated cleavage of cytoskeletal components. Caspase-3 cleaves ROCK1, generating a constitutively active fragment that induces hyperphosphorylation of myosin light chain, leading to actomyosin contraction and membrane blebbing [20]. Additionally, cleavage of gelsolin by caspase-3 produces an active fragment that severs actin filaments, contributing to cytoskeletal collapse [17]. Other structural proteins targeted include fodrin, paxillin, and focal adhesion kinases, which disrupts cell-matrix and cell-cell contacts, facilitating cell detachment and rounding [19].
The following diagram illustrates the key molecular events in Phase IIb apoptosis:
Experimental Models and Detection Methodologies
Induction Models for Late Apoptosis
Researchers employ various models to induce and study late apoptosis. Doxorubicin treatment (typically 5 μmol/L for HeLa cells) effectively triggers the intrinsic apoptotic pathway by intercalating into DNA and inhibiting topoisomerase II, causing DNA double-strand breaks and p53 activation [20]. The extrinsic pathway can be activated using death receptor ligands such as Fas ligand or TNF-related apoptosis-inducing ligand (TRAIL) at concentrations ranging from 10-100 ng/mL, depending on cell sensitivity [6] [21]. For cellular stress induction, reactive oxygen species inducers like hydrogen peroxide (100-500 μmol/L) or compounds that disrupt mitochondrial membrane potential are frequently utilized [22].
Detection and Imaging Techniques
Multiple complementary approaches are employed to detect and quantify Phase IIb apoptotic features:
Microscopy Techniques: Full-Field Optical Coherence Tomography (FF-OCT) provides label-free, high-resolution (sub-micrometer) visualization of apoptotic morphological changes, including membrane blebbing and apoptotic body formation in living cells [20]. Electron microscopy remains the gold standard for detailed ultrastructural analysis of nuclear condensation and organelle packaging in apoptotic bodies [17]. Fluorescence microscopy using DNA-binding dyes (DAPI, Hoechst) reveals nuclear fragmentation, while Annexin V conjugates detect phosphatidylserine externalization [17] [19].
Biochemical Assays: DNA laddering analysis via agarose gel electrophoresis detects the characteristic internucleosomal DNA cleavage pattern [19]. The TUNEL assay (Terminal deoxynucleotidyl transferase dUTP Nick-End Labeling) enzymatically labels the 3'-ends of DNA fragments, allowing in situ detection and quantification of DNA fragmentation [17]. Caspase activity assays using fluorogenic or colorimetric substrates confirm the activation of the executioner caspases, particularly caspase-3 [19].
The following workflow outlines a comprehensive experimental approach for studying late apoptosis:
Research Reagent Solutions
Table 3: Essential Reagents for Studying Phase IIb Apoptosis
| Reagent Category | Specific Examples | Application/Function | Detection Method |
|---|---|---|---|
| Apoptosis Inducers | Doxorubicin (5 μmol/L), Anti-Fas antibody, TRAIL (10-100 ng/mL), Staurosporine (0.1-1 μmol/L) | Trigger specific apoptotic pathways | Viability assays, morphology analysis |
| Caspase Substrates | Ac-DEVD-AMC (caspase-3), Ac-IETD-AFC (caspase-8), Ac-LEHD-AFC (caspase-9) | Measure caspase activity | Fluorometry, spectrophotometry |
| Nuclear Stains | DAPI, Hoechst 33342, Propidium Iodide | Visualize chromatin condensation and nuclear fragmentation | Fluorescence microscopy |
| Phosphatidylserine Detection | FITC-Annexin V, Cy5-Annexin V | Detect PS externalization on apoptotic bodies | Flow cytometry, microscopy |
| DNA Fragmentation Kits | TUNEL assay kits, DNA laddering extraction kits | Detect and quantify DNA breakdown | Fluorescence microscopy, gel electrophoresis |
| Caspase Inhibitors | Z-VAD-FMK (pan-caspase), Z-DEVD-FMK (caspase-3) | Confirm caspase-dependent mechanisms | Control experiments |
| Structural Protein Antibodies | Anti-lamin A/C, Anti-PARP, Anti-ROCK1 | Detect cleavage of specific substrates | Western blot, immunofluorescence |
Technical Considerations and Research Applications
Methodological Considerations
When studying Phase IIb apoptosis, several technical considerations are crucial for accurate interpretation. Kinetic monitoring is essential, as late apoptotic events represent a transient state that quickly progresses to secondary necrosis if apoptotic bodies are not cleared [20]. Employing multiple complementary detection methods is recommended, as reliance on a single parameter may yield false positives or negatives; for instance, Annexin V staining alone cannot distinguish between early and late apoptosis [17] [19].
The cell type and apoptotic stimulus significantly influence the morphological presentation and timing of late apoptotic events [20]. Additionally, researchers must consider that phagocytic clearance of apoptotic bodies occurs rapidly in vivo, making their detection more challenging in physiological contexts compared to in vitro systems [6] [13].
Research and Therapeutic Applications
Understanding Phase IIb apoptosis has significant implications for both basic research and therapeutic development. In drug discovery and screening, compounds that induce or enhance late apoptotic events are valuable candidates for cancer therapeutics, particularly for tumors resistant to conventional treatments [13] [19]. Quantitative assessment of nuclear fragmentation and apoptotic body formation serves as a key efficacy metric for evaluating novel chemotherapeutic agents [20] [23].
In toxicology and safety assessment, unintended induction of late apoptosis indicates compound toxicity, informing risk assessment [20]. Furthermore, dysregulated apoptotic body clearance is implicated in autoimmune and inflammatory diseases, making the morphological assessment of late apoptosis relevant for understanding disease mechanisms [6] [13]. The distinctive morphological features also provide important diagnostic markers in histopathology for distinguishing apoptotic cells from those undergoing other forms of cell death [17].
The Critical Role of Phagocytosis in Clearing Apoptotic Bodies without Inflammation
The efficient clearance of apoptotic cells is a fundamental biological process essential for maintaining tissue homeostasis and preventing inflammatory responses. This intricate process, known as efferocytosis, involves specialized mechanisms that allow phagocytes to recognize, engulf, and process dying cells without triggering the release of pro-inflammatory mediators. Understanding the molecular pathways governing this silent disposal system provides crucial insights into tissue remodeling, resolution of inflammation, and the prevention of autoimmune disorders. This technical review examines the sophisticated cellular and molecular machinery that enables the non-inflammatory clearance of apoptotic bodies, with particular emphasis on the morphological transitions during apoptosis and their implications for phagocytic recognition.
Apoptosis, or programmed cell death, is a genetically controlled process that eliminates unwanted or damaged cells during development, tissue homeostasis, and immune responses. In adult humans, an estimated one million cells undergo apoptosis every second, requiring an efficient clearance mechanism to prevent the accumulation of cellular debris [24]. The specific phagocytosis of dying cells by macrophages, termed efferocytosis, represents a critical mechanism for maintaining tissue integrity and preventing autoimmune reactions [25]. Unlike necrotic cell death, which results in membrane rupture and release of inflammatory contents, apoptosis produces membrane-bound fragments known as apoptotic bodies that are safely disposed of through phagocytic uptake.
The immunological consequences of apoptotic cell clearance are profoundly different from those following pathogen phagocytosis. While both processes may engage similar receptors, efferocytosis typically promotes an anti-inflammatory response and immunological tolerance rather than inflammation and immunity [26]. This review systematically examines the morphological features of apoptotic cells, the molecular recognition systems, and the intracellular processing mechanisms that collectively enable the non-inflammatory clearance of apoptotic bodies, with specific focus on their implications for research and therapeutic development.
Morphological Transitions During Apoptosis and Phagocytic Implications
The morphological progression of apoptosis creates distinct cellular states that directly influence how phagocytes recognize and engulf dying cells. These structural changes have been categorized into three sequential phases, each characterized by specific alterations that facilitate efficient clearance.
Table 1: Morphological Characteristics of Apoptotic Phases and Their Impact on Clearance
| Apoptotic Phase | Key Morphological Features | Detection Methods | Impact on Phagocytic Clearance |
|---|---|---|---|
| Phase I | Cell shrinkage, dense cytoplasm, decreased water content, increased eosinophilia, disappearance of microvilli | Electron microscopy, membrane permeability assays | Initial "find-me" signal release, early recognition marker exposure |
| Phase IIa | Chromatin condensation (pyknosis), marginalization along nuclear membrane, nuclear fragmentation | Fluorescence microscopy (Hoechst, DAPI, AO), TUNEL assay | Exposure of "eat-me" signals including phosphatidylserine |
| Phase IIb | Membrane blebbing, cytoskeleton degradation, apoptotic body formation | Light microscopy (HE, Giemsa, Wright's staining), FCM | Generation of bite-sized fragments for phagocytosis, maximal "eat-me" signal display |
Phase I: Initial Activation and Cell Shrinkage
During Phase I, apoptotic cells undergo cytoplasmic condensation and reduced volume while maintaining membrane integrity. The cytoplasm becomes increasingly dense with organelle compaction, and surface structures such as microvilli retract [14]. These changes are mediated by caspase activation and cytoskeletal reorganization. From a clearance perspective, this phase is characterized by the initial release of soluble "find-me" signals including nucleotides (ATP and UTP) and lipids (lysophosphatidylcholine) that attract potential phagocytes to the dying cell [25] [24].
Phase IIa: Nuclear Fragmentation
Phase IIa is defined by nuclear disintegration featuring highly condensed chromatin masses (pyknosis) that subsequently marginalize along the inner nuclear membrane [14]. This stage involves the activation of endogenous endonucleases that cleave DNA at internucleosomal sites, producing characteristic fragments of 180-200 base pairs [14]. Detection methods for this phase include fluorescence microscopy with DNA-binding dyes (Hoechst 33342, DAPI) that reveal chromatin condensation, and TUNEL assays that identify DNA strand breaks with 3'-OH ends [14]. The nuclear breakdown during Phase IIa coincides with the externalization of phosphatidylserine (PS) on the outer leaflet of the plasma membrane, serving as a primary "eat-me" signal for phagocytes [26].
Phase IIb: Apoptotic Body Formation
The terminal Phase IIb features extensive membrane blebbing and the formation of apoptotic bodies containing nuclear debris, organelles, and cytoplasmic components [14]. This process is mediated by ROCK1 (Rho-associated protein kinase 1) activation following caspase-3 cleavage, leading to actomyosin contraction and membrane protrusion [27]. The resulting apoptotic bodies range from 50-5000 nm in diameter and present an optimal "bite-size" for phagocytic engulfment [27]. These membrane-bound vesicles display the full complement of "eat-me" signals, including surface-exposed phosphatidylserine, calreticulin, and other apoptotic cell-associated molecular patterns (ACAMPs) that facilitate recognition by professional phagocytes [26] [28].
Diagram 1: Morphological Transitions During Apoptosis and Phagocytic Clearance. This flowchart illustrates the sequential phases of apoptosis and their relationship to phagocytic recognition mechanisms.
Molecular Recognition Systems in Efferocytosis
The precise recognition of apoptotic cells involves a sophisticated network of signaling molecules, receptors, and bridging proteins that distinguish dying cells from their viable counterparts. This molecular machinery ensures the selective removal of apoptotic bodies while maintaining immune silence.
Find-Me Signals and Phagocyte Recruitment
Apoptotic cells release chemoattractant signals that recruit potential phagocytes before the loss of membrane integrity. These "find-me" signals include:
- Nucleotides (ATP and UTP): Released through pannexin 1 channels and detected by phagocyte P2Y2 receptors [25] [24]
- Lysophosphatidylcholine (LPC): Generated by caspase-3 activation of calcium-independent phospholipase A2 (iPLA2) and recognized by G2A receptors on macrophages [25] [24]
- Sphingosine-1-phosphate (S1P): Promotes monocyte and macrophage migration through G protein-coupled receptors [25]
- Fractalkine (CX3CL1): Present on apoptotic microparticles and engages CX3CR1 on phagocytes [24]
These find-me signals operate at picomolar to nanomolar concentrations and establish a chemotactic gradient that guides phagocytes to apoptotic cells without provoking inflammatory responses [24].
Eat-Me Signals and Phagocytic Receptors
The specific recognition of apoptotic cells is mediated through "eat-me" signals that are absent from viable cells. The most characterized eat-me signal is phosphatidylserine (PS), a phospholipid normally restricted to the inner leaflet of the plasma membrane by ATP-dependent translocases [26]. During apoptosis, PS becomes externalized through caspase-activated scramblase activity and translocase inhibition [26]. Phagocytes recognize PS through multiple receptor systems:
Table 2: Principal Phagocytic Receptors and Recognition Mechanisms in Efferocytosis
| Receptor Category | Specific Receptors | Recognition Mechanism | Signaling Pathway |
|---|---|---|---|
| Direct PS Receptors | TIM-1, TIM-4, BAI1, Stabilin-2 | Direct binding to phosphatidylserine | BAI1: ELMO1-Dock180-Rac module; TIM-4: Requires adaptor for signaling |
| Bridging Molecule Receptors | Integrins (αvβ3, αvβ5), TAM receptors (Tyro3, Axl, Mer) | Bind opsonins (MFG-E8, Gas6, Protein S) that recognize PS | Integrins: Focal adhesion kinase; TAM receptors: Tyrosine kinase signaling |
| Scavenger Receptors | CD36, SR-A, LOX-1 | Recognize oxidized PS and other modified lipids | Src family kinases, Rho GTPases |
| Complement Receptors | CD91 (LRP1), C1q receptors | Bind complement proteins (C1q, C3b) that opsonize apoptotic cells | MAPK/ERK signaling |
The TAM receptor family (Tyro3, Axl, Mer) represents a particularly important recognition system that binds the bridging molecules Gas6 and Protein S, which in turn interact with PS on apoptotic cells [25]. Similarly, MFG-E8 (lactadherin) forms a molecular bridge between PS and phagocyte integrins αvβ3 and αvβ5 [25]. The developmental endothelial locus-1 (DEL-1) also facilitates efferocytosis by binding both PS and αvβ3 integrin, with compartmentalized expression determining its anti-inflammatory versus pro-resolution functions [25].
Don't-Eat-Me Signals
Viable cells express surface molecules that actively inhibit phagocytic engulfment. The best-characterized "don't-eat-me" signal is CD47, which engages signal regulatory protein alpha (SIRPα) on phagocytes to suppress engulfment [25]. During apoptosis, CD47 expression decreases, thereby removing this inhibitory signal [25]. Other don't-eat-me signals include CD31 (PECAM-1), which engages in homotypic interactions between viable cells and phagocytes to prevent inappropriate clearance [25].
Diagram 2: Molecular Recognition Systems in Efferocytosis. This diagram illustrates the key signaling pathways and molecular interactions between apoptotic cells and phagocytes during efferocytosis.
Intracellular Processing and Immunological Consequences
Following recognition and engulfment, the internalized apoptotic bodies undergo intracellular processing that shapes the subsequent immune response. The metabolic and signaling pathways activated during this process are critical for maintaining non-inflammatory clearance.
Engulfment and Phagolysosomal Degradation
The internalization of apoptotic bodies occurs through unique engulfment synapses that spatially organize recognition receptors and signaling components [25]. Unlike Fc receptor-mediated phagocytosis, which typically produces pro-inflammatory responses, efferocytosis triggers distinct signaling cascades that promote immune tolerance. The internalized apoptotic material is trafficked through the endocytic pathway and ultimately degraded in phagolysosomes, with the resulting metabolic byproducts influencing macrophage function [25].
Immunometabolic Reprogramming
Efferocytosis induces significant metabolic reprogramming in phagocytes that supports their anti-inflammatory phenotype. The digestion of apoptotic cell-derived membranes delivers a substantial lipid load that promotes fatty acid oxidation and mitochondrial respiration [25]. This metabolic shift away from glycolysis supports the production of anti-inflammatory mediators while limiting pro-inflammatory responses. Additionally, efferocytic macrophages upregulate enzymes such as 12/15-lipoxygenase that generate specialized pro-resolving mediators (SPMs) including resolvins, lipoxins, and maresins [25].
Anti-Inflammatory Mediator Production
The processing of apoptotic cells directly stimulates the production of immunosuppressive cytokines including TGF-β and IL-10 [25] [28]. These cytokines suppress the production of pro-inflammatory factors such as TNF, IL-1β, and IL-8, while promoting tissue repair and regeneration [25]. The strategic location of efferocytic receptors also contributes to immune silencing, as their engagement typically activates negative regulators of inflammatory signaling such as SOCS1 and SOCS3 [28].
Experimental Methods for Studying Apoptotic Cell Clearance
The investigation of efferocytosis requires specialized methodologies that can accurately quantify clearance efficiency and characterize the underlying molecular mechanisms. The following experimental approaches represent current best practices in the field.
In Vitro Co-culture Phagocytosis Assays
Live co-culture systems using fluorescently labeled apoptotic cells and macrophages enable real-time quantification of efferocytosis. The established protocol involves:
- Macrophage differentiation: THP-1 monocytes are differentiated into macrophages using 50 nM PMA (phorbol 12-myristate 13-acetate) for 48 hours [29]
- Apoptotic cell preparation: Target cells (e.g., prostate cancer PC3 or CL-1 cells) are engineered to express DsRed Express fluorescent protein or labeled with Cell Tracker Red CMTPX dye [29]
- Co-culture establishment: Fluorescent apoptotic cells are co-cultured with macrophages at optimized ratios (typically 1:10 to 1:20) in serum-free medium [29]
- Quantification: Phagocytosis is assessed using Confocal and Nomarski microscopy, with quantification of internalized fluorescent cells per macrophage [29]
This approach provides sensitive, measurable, and reproducible assessment of phagocytic activity that can be adapted for high-throughput screening of efferocytosis modulators.
Apoptosis Detection Methods
Different stages of apoptosis require specific detection strategies based on characteristic morphological and biochemical changes:
- Early apoptosis: Analysis of mitochondrial membrane potential using fluorescent cationic dyes (e.g., JC-1, TMRM) that exhibit potential-dependent accumulation in mitochondria [14]
- Mid-stage apoptosis: Annexin V staining for phosphatidylserine exposure combined with viability dyes (e.g., propidium iodide) to distinguish from necrotic cells [14] [28]
- Late apoptosis: DNA fragmentation detection via TUNEL assay or gel electrophoresis for nucleosomal ladder patterns [14]
- Morphological assessment: Electron microscopy for ultrastructural changes across all phases; light microscopy for Phase IIb apoptotic bodies [14]
Apoptotic Body Isolation and Characterization
The isolation and analysis of apoptotic bodies requires specialized techniques due to their heterogeneous size and composition:
- Differential centrifugation: Sequential centrifugation at increasing speeds (2,000 × g for 10 min followed by 10,000 × g for 30 min) to separate ABs from larger debris and smaller vesicles [27]
- Fluorescence-activated cell sorting (FACS): Isolation of ABs based on specific surface markers (phosphatidylserine, caspase-3, calreticulin) and light scattering properties [27]
- Characterization methods: Nanoparticle tracking analysis (NTA) for size distribution, cryo-electron microscopy for morphological assessment, and Western blotting for protein marker confirmation [27]
Table 3: Key Methodologies for Apoptotic Cell Clearance Research
| Experimental Goal | Recommended Methods | Key Readouts | Technical Considerations |
|---|---|---|---|
| Phagocytosis Quantification | Live co-culture with fluorescent targets, time-lapse imaging | Internalized targets per phagocyte, phagocytic index | Requires differential staining to distinguish attached vs. internalized targets |
| "Eat-Me" Signal Detection | Annexin V staining, antibody labeling for oxidized lipids | Surface PS exposure, oxidized epitope presentation | Must confirm apoptosis specificity with caspase inhibition |
| Phagocyte Recruitment | Transwell migration assays, microfluidic devices | Phagocyte migration toward apoptotic conditioned media | Distinguish chemotaxis from chemokinesis through checkerboard analysis |
| In Vivo Clearance Assessment | Intravital microscopy, labeled apoptotic cell injection | Clearance kinetics, phagocyte recruitment in tissue context | Consider anatomical differences in clearance efficiency |
The Scientist's Toolkit: Essential Research Reagents
The following table compiles critical reagents and their applications for investigating apoptotic cell clearance mechanisms, derived from current methodological approaches.
Table 4: Essential Research Reagents for Apoptotic Clearance Studies
| Reagent Category | Specific Examples | Research Application | Mechanistic Role |
|---|---|---|---|
| Phagocyte Modulators | PMA (phorbol 12-myristate 13-acetate), Pigment Epithelium-Derived Factor (PEDF) | Macrophage differentiation, phagocytosis enhancement | PKC activation, efferocytosis potentiation |
| Fluorescent Labels | DsRed Express, Cell Tracker Red CMTPX, Annexin V-FITC | Target cell labeling, PS exposure detection | Phagocytosis quantification, apoptosis staging |
| Receptor Blockers | Anti-TIM-4, anti-αvβ3 integrin, anti-CD36 antibodies | Pathway-specific inhibition | Mechanistic dissection of recognition systems |
| "Find-Me" Signal Receptors | P2Y2 agonists (ATP, UTP), G2A ligands (LPC) | Phagocyte recruitment studies | Chemotaxis assessment, signal transduction analysis |
| Metabolic Inhibitors | Fatty acid oxidation inhibitors, 12/15-LOX inhibitors | Immunometabolic studies | Resolution mediator production, metabolic reprogramming |
| Apoptosis Inducers | Staurosporine, actinomycin D, TNF-α/CHX | Controlled apoptosis induction | Standardized apoptotic cell preparation |
The non-inflammatory clearance of apoptotic bodies represents a sophisticated biological system that maintains tissue homeostasis while preventing inappropriate immune activation. The process integrates specific morphological changes during apoptosis with specialized recognition mechanisms and intracellular processing pathways that collectively ensure silent disposal of dying cells. Understanding these mechanisms provides not only fundamental biological insights but also therapeutic opportunities for manipulating efferocytosis in disease contexts ranging from chronic inflammation to autoimmunity. Continued technical innovation in tracking, quantifying, and modulating apoptotic cell clearance will further illuminate this critical biological process and its translational applications.
Detection and Analysis: Techniques for Visualizing Apoptotic Morphology
Apoptosis, or programmed cell death, is a fundamental process crucial for maintaining tissue homeostasis, organ development, and the elimination of damaged or mutant cells [14]. Its detection and accurate quantification are therefore paramount in both basic research and applied drug development. While numerous biochemical and molecular techniques exist, the morphological assessment of apoptosis remains a cornerstone, providing direct and often unequivocal evidence of cell death within its tissue context [30]. This technical guide focuses on the application of common light microscopy stains—Hematoxylin and Eosin (H&E), Giemsa, and Wright's—for identifying apoptotic cells, with its content rigorously framed within the classical morphological model that defines three phases of apoptosis: Phase I (cell shrinkage), Phase IIa (nuclear condensation), and Phase IIb (apoptotic body formation) [14].
The significance of morphology was cemented by Kerr, Wyllie, and Currie in 1972, who coined the term "apoptosis" to describe a specific morphological pattern of cell death distinct from necrosis [31] [30]. Despite the advent of sophisticated assays, guidelines advise that morphological characteristics are the definitive arbiter for diagnosing apoptosis, as biochemical features like DNA fragmentation can vary by cell type and lead to false negatives [30]. Light microscopy, with its simplicity, cost-effectiveness, and capacity for providing storable specimens for further study, is an accessible and powerful tool for this purpose [14] [32]. This guide will detail how H&E, Giemsa, and Wright's staining can be deployed to detect the hallmark morphological features across the different phases of apoptosis, providing researchers and drug development professionals with clear methodologies and interpretive frameworks.
Morphological Hallmarks of Apoptosis Across Three Phases
The progression of apoptosis is characterized by a sequence of distinct structural changes. The following table summarizes the key features observable via light microscopy, correlated with the three morphological phases [14].
Table 1: Morphological Features of Apoptosis Across Key Phases
| Phase | Morphological Feature | Description |
|---|---|---|
| Phase I | Cell Shrinkage | Decreased cell volume, dense cytoplasm, loss of cell-cell contact [14] [30]. |
| Phase IIa | Chromatin Condensation | Nuclear chromatin condenses into dense masses (pyknosis) or marginalizes along the inner nuclear membrane [14]. |
| Phase IIb | Apoptotic Body Formation | The cell membrane buds off, forming small, membrane-bound vesicles containing cytoplasm and nuclear debris [14] [33]. |
These features form the basis for identifying apoptotic cells under the light microscope. It is important to note that cell shrinkage is one of the most ubiquitous characteristics, occurring in almost all instances of apoptosis regardless of the initiating stimulus [30]. Furthermore, the formation of apoptotic bodies is considered an important morphological marker, and their rapid phagocytosis by neighboring cells in vivo prevents inflammatory responses, making their detection in a small area challenging [14].
Staining Techniques and Their Applications
The choice of stain directly influences the ease and clarity with which these morphological features can be visualized. The following table provides a comparative overview of H&E, Giemsa, and Wright's staining for apoptosis detection.
Table 2: Comparison of Staining Methods for Apoptosis Detection by Light Microscopy
| Staining Method | Staining Principle | Key Apoptotic Features Visualized | Advantages & Disadvantages | Optimal Apoptosis Phase for Detection |
|---|---|---|---|---|
| H&E | Hematoxylin (basic) stains nucleic acids blue; Eosin (acidic) stains proteins pink [30]. | Cell shrinkage, cytoplasmic eosinophilia, nuclear pyknosis, and apoptotic bodies [14] [32]. | Advantages: Routine, widely available, provides good tissue context [32].Disadvantages: Chromatic differentiation can limit easy identification; may underestimate apoptosis [32]. | Phase IIb (apoptotic bodies) [14]. |
| Giemsa | Romanowsky-type stain; azure dyes and eosin differentiate cellular components. | Cell shrinkage, chromatin condensation, and apoptotic body formation [34] [33]. | Advantages: Excellent for highlighting nuclear detail and morphology in cytospin preparations [30].Disadvantages: Requires consistent protocol for reproducible results. | Phase IIa and IIb [33]. |
| Wright's | Similar to Giemsa; a Romanowsky stain based on methylene blue and eosin. | Cell shrinkage, chromatin condensation, and loss of surface microvilli [35] [33]. | Advantages: Standard in hematology; ideal for blood smears and suspended cells [35] [36].Disadvantages: Staining procedure requires preparation time and infrastructure [35]. | Phase IIa and IIb [33]. |
Experimental Protocols for Key Stains
Protocol for Giemsa Staining [34] [33]
- Sample Preparation: Plate cells (e.g., PC-3 or CEM-SS leukemia cells) on a slide, for instance, via cytospin centrifugation.
- Fixation: Fix cells with 75% methanol for 10 minutes at room temperature.
- Staining: Apply Giemsa working solution (diluted with phosphate buffer as per supplier's instructions) for the prescribed duration.
- Washing: Gently rinse the slide with distilled water to remove excess stain.
- Air-Drying: Allow the slide to air-dry completely.
- Observation: Examine under a light microscope using a 200x or higher magnification objective.
Protocol for H&E Staining [32] [30]
- Sample Preparation: Deparaffinize and rehydrate formalin-fixed, paraffin-embedded tissue sections through a graded alcohol series.
- Nuclear Staining: Immerse slides in Hematoxylin solution for a specified time (e.g., 5-10 minutes).
- Washing: Rinse in running tap water.
- Differentiation: Briefly dip in 1% acid alcohol (1% HCl in 70% ethanol).
- Bluing: Place in Scott's tap water or a weak ammonia solution.
- Cytoplasmic Staining: Counterstain with Eosin Y working solution (0.25-1%) for 1-5 minutes.
- Dehydration & Mounting: Dehydrate through graded alcohols, clear in xylene, and mount with a synthetic resin.
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 3: Key Reagent Solutions for Morphological Apoptosis Detection
| Reagent / Solution | Function / Application in Apoptosis Detection |
|---|---|
| Hematoxylin | A basic dye that binds to DNA/RNA, staining the nucleus blue-purple, allowing visualization of chromatin condensation and nuclear fragmentation [30]. |
| Eosin Y | An acidic dye that binds to cationic amino groups in proteins, staining the cytoplasm pink-red, highlighting cytoplasmic condensation and increased eosinophilia [30]. |
| Giemsa Stain | A compound stain that differentially colors cellular components; particularly effective for visualizing condensed chromatin in apoptotic nuclei [34] [33]. |
| Wright's Stain | A hematology stain used to distinguish blood cell types; reveals cell shrinkage and chromatin changes in apoptotic leukocytes [35] [33]. |
| Methanol | Common fixative used to preserve cellular morphology prior to staining (e.g., in Giemsa protocol) [34]. |
| Phosphate Buffered Saline (PBS) | An isotonic solution used for washing cells and preparing reagent dilutions to maintain physiological pH and osmolarity [30]. |
Integrating Morphology with Biochemical Pathways
The morphological changes observed with H&E, Giemsa, and Wright's stains are the phenotypic endpoint of complex, regulated biochemical pathways. Apoptosis can be triggered via extrinsic (death receptor) or intrinsic (mitochondrial) pathways. The intrinsic pathway, outlined below, is particularly relevant to the morphological phases, as it leads to the activation of caspases and endonucleases that execute the cellular dismantling process [14].
Diagram 1: Intrinsic Apoptosis Pathway Leading to Morphological Changes. This pathway illustrates how an apoptotic stimulus triggers mitochondrial events, culminating in the caspase and endonuclease activation that drives the morphological phases detectable by light microscopy.
Light microscopy using H&E, Giemsa, and Wright's staining provides a foundational and purpose-dependent approach for identifying apoptotic cells [14]. Each stain offers unique advantages: H&E is the universal standard for histology, providing excellent tissue context; Giemsa offers superb nuclear detail; and Wright's is indispensable for hematological applications. Their effective use requires a deep understanding of the underlying morphological phases of apoptosis—shrinkage, condensation, and fragmentation—as these features form the basis for identification and interpretation.
For researchers in drug development, these staining methods are a first-line tool for rapidly screening the cytotoxic or cytostatic effects of novel compounds, as demonstrated in studies with anticancer anthraquinones on leukemia cells [33]. However, it is critical to acknowledge the limitations. Morphological analysis can be subjective and may miss early-stage apoptotic cells. Furthermore, the complexity of cell death mechanisms means that features of different death pathways (e.g., apoptosis and autophagy) can sometimes overlap [30]. Therefore, for conclusive evidence, morphological assessment should be combined with other biochemical or molecular techniques, such as TUNEL assay for DNA fragmentation or caspase activation assays, to create a comprehensive and irrefutable picture of apoptotic cell death [14] [37]. This multi-faceted approach ensures accurate data, which is crucial for both basic scientific discovery and the development of new therapeutics.
Transmission Electron Microscopy (TEM) stands as a critical technique in cell biology for the detailed visualization of apoptotic processes. Apoptosis, or programmed cell death, is a tightly regulated mechanism vital for tissue differentiation, organ development, aging, and the elimination of damaged or mutant cells [14]. The ability to precisely identify and characterize the morphological stages of apoptosis is fundamental to research in cancer, neurodegenerative diseases, and drug development. Unlike other methods that may only detect specific biochemical events, TEM provides unparalleled resolution of the ultrastructural changes that define the entire apoptotic cascade, from initial induction to phagocytic clearance [38] [39]. This technical guide details the application of TEM for revealing these defining features across all phases of apoptosis, contextualized within a broader morphological research framework.
The Morphological Phases of Apoptosis as Visualized by TEM
The progression of apoptosis is categorized into distinct phases based on characteristic morphological alterations in the nucleus and cytoplasm, which TEM is uniquely positioned to elucidate.
Phase I: Early Apoptotic Changes
The initial phase is marked by cell commitment to death. TEM reveals several key ultrastructural changes:
- Cell Shrinkage: The cell undergoes a reduction in volume, acquiring a dense cytoplasm [14].
- Loss of Surface Specializations: Structures like microvilli on the cell surface disappear [14].
- Organelle Alterations: The endoplasmic reticulum often dilates, and numerous vacuoles, referred to as cavitations, become apparent within the cytoplasm [14].
- Chromatin Condensation: The chromatin begins to condense, though the nuclear membrane remains intact [14].
Phase IIa: Nuclear Fragmentation
This phase is dominated by decisive changes in the nucleus.
- Chromatin Agglutination and Marginalization: The chromatin becomes highly condensed and aggregates into dense masses (pyknosis) or assembles along the inner nuclear membrane [14].
- Nuclear Fragmentation: The nucleus breaks apart into discrete, membrane-bound fragments [14].
Phase IIb: Apoptotic Body Formation
The final stage of cellular disintegration involves:
- Membrane Blebbing: The cell membrane develops protrusions and invaginations [14] [39].
- Formation of Apoptotic Bodies: The cell fragments into multiple, membrane-bound vesicles known as apoptotic bodies. These bodies contain nuclear debris, intact organelles, and cytoplasmic components [14] [39]. The formation of apoptotic bodies is a key morphological marker of apoptosis [14].
Resolution: Phagocytosis
Apoptosis concludes with the efficient clearance of apoptotic cells without provoking an inflammatory response. TEM can show apoptotic bodies being recognized and engulfed by neighboring phagocytic cells, a process facilitated by "eat-me" signals like phosphatidylserine on the apoptotic body surface [39].
Table 1: Ultrastructural Changes in Apoptosis Phases Observed via TEM
| Apoptotic Phase | Nuclear Morphology | Cytoplasmic & Organelle Changes | Overall Cell Structure |
|---|---|---|---|
| Phase I | Chromatin begins to condense | Cell shrinkage; concentrated cytoplasm; dilation of endoplasmic reticulum; appearance of vacuoles (cavitations); loss of microvilli [14] [39] | Cell detaches from neighbors; membrane integrity maintained [14] |
| Phase IIa | Highly condensed, marginalized chromatin; nuclear fragmentation (pyknosis) [14] | - | Cell remains intact, no release of cellular contents [14] |
| Phase IIb | Nuclear fragments packaged into apoptotic bodies | Degradation of cytoskeleton; formation of membrane-bound apoptotic bodies containing cytoplasm, organelles, and nuclear debris [14] [39] | - |
| Resolution | - | - | Phagocytosis of apoptotic bodies by neighboring cells; no inflammation [39] |
TEM Experimental Protocol for Apoptosis Detection
A standardized protocol is essential for reliably capturing the ultrastructural landscape of apoptosis.
Sample Preparation (Primary Fixation to Embedding)
- Primary Fixation: Immediately after collection, immerse tissue samples (≈1 mm³) or pelleted cells in a buffered aldehyde fixative (e.g., 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer, pH 7.4) for a minimum of 2 hours at 4°C. This crosslinks proteins and preserves structure.
- Washing: Rinse samples thoroughly several times in the same buffer (0.1M sodium cacodylate) to remove excess fixative.
- Post-Fixation: Treat samples with 1% osmium tetroxide in buffer for 1-2 hours at 4°C. Osmium tetroxide stabilizes lipids and provides electron density to membranes.
- Dehydration: Gradually dehydrate the samples using a graded series of ethanol or acetone (e.g., 50%, 70%, 90%, 100%) to remove all water.
- Infiltration and Embedding: Infiltrate the samples with a resin, such as EPON or Araldite, beginning with a 1:1 mixture of resin and solvent, then pure resin. Finally, embed the samples in fresh resin and polymerize in an oven at 60°C for 24-48 hours.
Sectioning, Staining, and Imaging
- Ultramicrotomy: Use an ultramicrotome to cut ultrathin sections (60-90 nm) from the polymerized resin block. Mount sections on copper or nickel TEM grids.
- Contrast Staining: Stain the grids with heavy metal stains to enhance contrast.
- TEM Imaging: Examine the stained grids using a Transmission Electron Microscope operated at an appropriate accelerating voltage (e.g., 80-100 kV). Systematically image cells at various magnifications to document the full spectrum of apoptotic features.
Diagram 1: TEM sample preparation workflow
Correlating TEM Findings with Apoptosis Pathways
The morphological changes observed by TEM are the physical manifestation of underlying biochemical pathways. The two primary pathways are the intrinsic (mitochondrial) and extrinsic (death receptor) pathways, which converge on a common execution phase [38] [39].
- The Intrinsic (Mitochondrial) Pathway: Triggered by internal stresses like DNA damage or oxidative stress, this pathway is regulated by the Bcl-2 family of proteins. A shift in balance towards pro-apoptotic proteins (e.g., Bax, Bak) leads to mitochondrial outer membrane permeabilization (MOMP), resulting in the release of cytochrome c and other factors into the cytosol [14] [39]. Cytochrome c, in conjunction with Apaf-1, forms the "apoptosome," which activates caspase-9, initiating the caspase cascade [38].
- The Extrinsic (Death Receptor) Pathway: Initiated by the binding of extracellular death ligands (e.g., FasL) to cell surface death receptors (e.g., Fas). This receptor-ligand interaction recruits adapter proteins to form the Death-Inducing Signaling Complex (DISC), leading to the activation of caspase-8 [38] [39].
- Execution Phase: Both pathways culminate in the activation of effector caspases, primarily caspase-3. These executioner caspases systematically cleave hundreds of cellular substrates, including structural proteins like nuclear lamins and cytoskeletal components, directly causing the characteristic morphological changes of apoptosis, such as chromatin condensation, nuclear fragmentation, and membrane blebbing [39].
Diagram 2: Apoptosis signaling pathways
The Scientist's Toolkit: Research Reagent Solutions
Successful TEM-based apoptosis research relies on specific reagents for inducing, processing, and identifying apoptotic cells.
Table 2: Key Reagents for Apoptosis and TEM Research
| Reagent/Category | Function & Role in Apoptosis Research | Example Application in Context |
|---|---|---|
| Chemical Inducers | Trigger apoptosis via specific pathways for experimental study. | Staurosporine (broad-spectrum kinase inhibitor) used at 0.5µM to induce intrinsic apoptosis in mesenchymal stem cells [40]. |
| Fixatives | Preserve cellular ultrastructure instantly by cross-linking proteins; critical for artifact-free TEM. | Primary fixation with 2.5% Glutaraldehyde; post-fixation with 1% Osmium Tetroxide to stabilize lipids and membranes [38]. |
| Heavy Metal Stains | Bind to cellular components to create electron contrast for TEM visualization. | Uranyl Acetate and Lead Citrate used sequentially to stain nucleic acids and membranes, revealing chromatin condensation and organelle details [14]. |
| Immunogold Labels | Enable highly specific localization of target proteins at the ultrastructural level via antibody-gold conjugates. | Antibodies against activated Caspase-3 or cytochrome C can be used to correlate protein presence/translocation with morphological phases. |
| Inhibitors | Block specific apoptotic pathways to study mechanism or confer protection. | zVAD-FMK (pan-caspase inhibitor) used in vivo at 1 mg/kg to inhibit apoptosis and assess functional outcomes in disease models [41]. |
Comparative Analysis: TEM vs. Other Apoptosis Detection Methods
While TEM is the gold standard for morphological confirmation, it is one tool in a broader methodological arsenal. Understanding its relative strengths and weaknesses guides appropriate experimental design.
Table 3: Comparing Apoptosis Detection Techniques
| Method | Key Principle | Advantages | Disadvantages / Limitations | Suitability for Apoptosis Phase |
|---|---|---|---|---|
| TEM | High-resolution imaging of ultrastructure. | Direct, intuitive observation of hallmark morphological features (e.g., chromatin condensation, apoptotic bodies); high spatial resolution [14] [38]. | End-point assay (non-viable cells); time-consuming sample prep; requires high skill level; potential for false positives; typically reveals later stages [14] [38]. | All phases (I, IIa, IIb), with Phase IIb being most obvious [14]. |
| Light Microscopy | Observation of stained cells for gross morphological changes. | Simplicity, convenience; storable specimens; reveals cell shrinkage and apoptotic bodies [14]. | Limited resolution; apoptosis in a small area is easily missed; mainly suitable for Phase IIb [14]. | Primarily Phase IIb [14]. |
| DNA Gel Electrophoresis | Detection of internucleosomal DNA cleavage (DNA laddering). | Simple, qualitatively accurate for DNA fragmentation [14]. | Poor specificity & sensitivity; semi-quantitative; cannot localize apoptotic cells; not suitable for early apoptosis [14]. | Middle to late stages [14]. |
| TUNEL Assay | Labeling of 3'-OH ends of DNA fragments in situ. | Relatively sensitive and specific; allows for counting and quantifying apoptotic cells in tissue sections [14]. | Can yield false-positive results (e.g., in necrotic cells); requires careful controls; suitable for late-stage apoptosis [14]. | Late-stage apoptosis [14]. |
| Flow Cytometry | Multiparametric analysis of single cells in suspension. | High-throughput quantitative data; can measure Annexin V/PI, caspase activation, mitochondrial membrane potential. | Cannot visualize ultrastructural morphology; requires single-cell suspensions. | Varies with the specific assay (early to late). |
| Mitochondrial Potential Probes | Fluorescent dyes detecting loss of mitochondrial inner membrane potential (ΔΨm). | Can detect an early marker of the intrinsic apoptotic pathway [14]. | Affected by changes in pH; requires careful calibration [14]. | Early stage (intrinsic pathway) [14]. |
Transmission Electron Microscopy remains an indispensable technique for the definitive identification and characterization of apoptosis, providing a direct window into the ultrastructural transformations that define Phases I, IIa, and IIb. Its power is maximized when integrated into a multidisciplinary approach, correlating high-resolution morphology with biochemical assays and functional data. As research continues to unveil novel regulators of cell death, such as recently identified molecular switches within the apoptotic machinery [42], and explores therapeutic modulation of apoptosis in disease models [41], TEM will continue to be a cornerstone technology for validating and contextualizing scientific discoveries in the field of programmed cell death.
The detection of apoptotic cells is a cornerstone of biomedical research, particularly in cancer biology and drug development. A defining feature of apoptosis is a series of characteristic morphological changes in the cell nucleus, including chromatin condensation and nuclear fragmentation. Fluorescence microscopy, especially confocal microscopy, serves as a powerful tool for visualizing these changes, largely through the use of specific nuclear stains like Hoechst, DAPI, and Acridine Orange (AO). When used within confocal systems, these dyes provide the high-resolution, optically sectioned images necessary to distinguish the subtle nuclear morphologies that characterize different phases of apoptotic cell death. This technical guide details the application of these vital dyes for nuclear assessment within the context of a broader thesis on the morphological features of apoptosis phases I, IIa, and IIb.
Technical Specifications of Nuclear Stains
The effective use of nuclear stains requires a thorough understanding of their photophysical properties, staining specificities, and optimal working conditions. The table below summarizes the key technical data for Hoechst, DAPI, and Acridine Orange to facilitate experimental planning and comparison.
Table 1: Technical Specifications and Staining Protocols for Hoechst, DAPI, and Acridine Orange
| Feature | Hoechst 33342 | DAPI | Acridine Orange (AO) |
|---|---|---|---|
| Primary Use | Nuclear counterstain; cell cycle & apoptosis studies [43] | Nuclear counterstain; apoptosis studies [14] | Viability assay; differential staining of DNA/RNA [14] |
| DNA Binding Mode | Minor-groove binder, A/T preference [44] | Minor-groove binder, A/T preference [44] | Intercalation and electrostatic interaction [14] |
| Excitation/Emission (nm) | 350/461 [43] | 358/461 [44] | Information missing from search results |
| Recommended Filter Set | DAPI [43] | DAPI | Information missing from search results |
| Cell Permeability | High (live cell preferred) [44] | Low (fixed cell preferred) [44] | High (live cell) |
| Recommended Staining Concentration | 1 µg/mL (live & fixed) [44] | 10 µg/mL (live); 1 µg/mL (fixed) [44] | Information missing from search results |
| Key Considerations | - Mutagen; handle with care [43]- Fluorescence quenched by BrdU [43]- Can induce apoptosis in some cell types [44] | - Less cell-permeant than Hoechst [44]- Can be added to mounting medium [44]- Subject to UV photoconversion [44] | - Emits different colors based on binding to DNA (green) or RNA (red) [14] |
Nuclear Morphology Across Apoptotic Phases
Apoptosis progresses through distinct phases, each with characteristic nuclear morphology that can be identified with nuclear stains [14]:
- Phase I: Cells shrink, and the cytoplasm becomes dense. While initial chromatin condensation begins, the most definitive nuclear changes are observed in later phases [14].
- Phase IIa: This phase features prominent chromatin condensation, where chromatin becomes highly condensed (pyknosis) or assembles along the inner nuclear membrane (chromatin margination). This is a key stage detectable with Hoechst, DAPI, and AO staining [14].
- Phase IIb: The nucleus breaks into fragments (karyorrhexis), and the cell forms membrane-coated apoptotic bodies containing nuclear debris and organelles [14].
These morphological changes are readily visible under fluorescence or confocal microscopy after staining with Hoechst 33342, DAPI, or AO, which reveal the intensity and distribution of fluorescence signals corresponding to the state of the nucleus and chromatin [14].
Experimental Protocols for Nuclear Staining
Staining Live Cells with Hoechst 33342
This protocol is optimized for staining live cells for subsequent imaging, such as in confocal microscopy.
You will need:
- Cells growing in an appropriate culture vessel
- Hoechst 33342 stock solution (e.g., 10 mg/mL in water) [43]
- Complete culture medium
- Phosphate-buffered saline (PBS) [43]
Procedure:
- Prepare Staining Solution: Dilute the Hoechst 33342 stock solution in pre-warmed complete culture medium to a final concentration of 1 µg/mL [44].
- Apply Solution: Remove the existing culture medium from the cells and replace it with a sufficient volume of the staining solution to cover the cells [43].
- Incubate: Incubate the cells for 5–15 minutes at room temperature or 37°C, protected from light [43] [44].
- Wash and Image: Remove the staining solution and wash the cells 3 times with PBS [43]. Image the cells directly in PBS or an appropriate imaging medium.
Protocol Tip: For a less disruptive process, you can add a 10X concentrated dye solution directly to the culture medium (e.g., 10 µL of 10 µg/mL Hoechst into 1 mL medium), mixing immediately and gently [44].
Staining Fixed Cells or Tissue Sections with DAPI
DAPI is generally preferred for fixed samples due to its lower cell permeability in live cells [44].
You will need:
- Fixed and permeabilized cells or tissue sections
- DAPI stock solution (e.g., 10 mg/mL in water) [44]
- Phosphate-buffered saline (PBS)
Procedure:
- Prepare Staining Solution: Dilute the DAPI stock solution in PBS to a final concentration of 1 µg/mL [44].
- Apply Solution: Add the DAPI/PBS solution to cover the fixed cells or tissue sections.
- Incubate: Incubate for at least 5 minutes at room temperature, protected from light [44].
- Image: The samples can be imaged immediately. Washing is optional but not required. For permanent mounting, DAPI can be included directly in an antifade mounting medium [44].
Workflow for Nuclear Staining and Apoptosis Assessment
The following diagram illustrates the critical decision points and steps in the experimental workflow for preparing and analyzing samples using these nuclear stains.
The Scientist's Toolkit: Key Research Reagent Solutions
Successful experimentation relies on high-quality reagents. The following table lists essential materials and their functions for experiments involving nuclear staining and confocal microscopy.
Table 2: Essential Research Reagents for Nuclear Staining and Confocal Microscopy
| Reagent/Material | Function/Application | Technical Notes |
|---|---|---|
| Hoechst 33342 | Cell-permeant nuclear counterstain for live/fixed cells; distinguishes condensed nuclei in apoptosis [43] [44] | Prepare 10 mg/mL stock in dH₂O; store at 2–6°C or ≤ -20°C [43]. Mutagen—handle with care [43]. |
| DAPI (4′,6-diamidino-2-phenylindole) | Nuclear counterstain, preferred for fixed cells; stains apoptotic nuclei and chromocenters [14] [44] [45] | Use DAPI dilactate for better solubility [44]. Fluorescence is quenched by BrdU [43]. |
| Acridine Orange (AO) | Metachromatic dye for cell viability; differentially stains DNA (green) and RNA (red) [14] | Allows for simultaneous assessment of nuclear morphology and RNA content in a single stain [14]. |
| Phosphate-Buffered Saline (PBS) | Physiological buffer for washing cells and diluting staining solutions [43] | Ensure pH is stable (e.g., 7.4) for consistent results. |
| Antifade Mounting Medium | Preserves fluorescence in fixed samples during microscopy and storage [44] | Available with or without DAPI for one-step mounting and staining [44]. |
| Confocal Microscope | High-resolution fluorescence imaging with optical sectioning to reject out-of-focus light [46] | Provides the resolution needed to distinguish fine apoptotic nuclear structures [14] [46]. |
Principles and Advantages of Confocal Microscopy
Confocal microscopy is indispensable for high-resolution imaging of apoptotic nuclei in thick samples. Its core principle involves focusing both illumination and detection optics on a single, diffraction-limited spot in the sample, using a pinhole aperture in front of the detector to reject out-of-focus light [46]. This optical sectioning capability allows for the clear visualization of nuclear details in one focal plane without the blurring haze that characterizes widefield microscopy [46]. Furthermore, by collecting a series of these optical sections (a z-stack), a high-resolution 3D reconstruction of the sample can be generated [46].
The lateral resolution of a confocal microscope is superior to a conventional widefield microscope and can be described by the formula:
R_lateral = 0.4λ / NA [46]
Where λ is the emission wavelength of the fluorophore (e.g., 461 nm for Hoechst/DAPI) and NA is the numerical aperture of the objective lens [46]. A higher NA objective provides better resolution. For dim samples, a trade-off exists between resolution and signal-to-noise; the pinhole can be opened to collect more light at the cost of some resolution [46].
Assessing Apoptotic Phases via Nuclear Morphology
The high contrast and resolution provided by confocal microscopy, combined with specific nuclear stains, enable researchers to identify and characterize the phases of apoptosis based on nuclear morphology. The following diagram summarizes the key morphological criteria and how they are assessed.
Critical Technical Considerations
- Dye Selection: The choice between Hoechst and DAPI is critical. Hoechst 33342 is generally preferred for live-cell imaging due to its superior cell permeability and lower immediate toxicity [44]. DAPI is an excellent and stable choice for fixed cells and can be conveniently added to mounting media [44].
- Artifacts and Limitations: Be aware that a "green haze" in images can indicate excessive Hoechst dye concentration [43]. Both DAPI and Hoechst are susceptible to photoconversion when exposed to UV light, causing them to fluoresce in other channels; this can be mitigated by imaging green fluorescence before UV exposure or using hardset mounting media [44].
- Microscope Configuration: To achieve the best resolution in confocal microscopy, use an objective lens with the highest possible Numerical Aperture (NA) and ensure the pinhole is set to 1 Airy unit or smaller [47] [46]. Remember that axial (z-axis) resolution is inherently lower than lateral resolution [46].
Advantages and Limitations of Each Morphological Detection Technique
Within the broader thesis on morphological features of apoptosis phases I, IIa, and IIb research, the selection of an appropriate detection technique is paramount. Apoptosis, or programmed cell death, is a fundamental process in tissue development, homeostasis, and the elimination of damaged cells. Its dysregulation is implicated in numerous diseases, including cancer and neurodegenerative disorders [14] [48]. Research into apoptotic pathways is thus critical for therapeutic development.
The morphological progression of apoptosis is categorized into three distinct phases. In Phase I, cells undergo shrinkage, cytoplasm condensation, and loss of microvilli. Phase IIa is characterized by nuclear changes, including chromatin condensation (pyknosis) and margination along the inner nuclear membrane. Finally, Phase IIb involves nuclear fragmentation (karyorrhexis) and the formation of membrane-bound apoptotic bodies [14]. Accurately identifying these phases requires techniques capable of capturing specific morphological hallmarks.
This technical guide provides an in-depth comparison of morphological detection techniques, evaluating their advantages, limitations, and optimal application within apoptosis research. It is designed to help researchers and drug development professionals select the most suitable method based on their specific experimental needs, phase of interest, and technical constraints.
Core Morphological Techniques for Apoptosis Detection
A range of techniques is available for detecting apoptosis based on morphological changes, each with unique capabilities for resolving the key features of different apoptotic phases.
Light Microscopy
- Methodology: Cells or tissue sections are stained with dyes such as Hematoxylin and Eosin (H&E), Giemsa, or Wright's stain. Stained samples are then visualized under a light microscope to identify classic apoptotic morphology [14].
- Detectable Features: This method is primarily suitable for observing Phase IIb apoptosis, allowing researchers to identify cell rounding, nuclear shedding, and the presence of apoptotic bodies [14]. Some surface markers on cell debris that facilitate phagocytosis can also be observed.
Electron Microscopy
- Methodology: Samples are stained with heavy metals like uranyl acetate and lead citrate. A transmission electron microscope (TEM) is then used to examine the ultra-structural details of cells at very high resolution [14].
- Detectable Features: TEM is the gold standard for detailed morphological analysis and is suitable for observing all phases of apoptosis (I, IIa, and IIb). It can reveal early Phase I changes like cytoplasmic vacuolation (cavitations) and cell shrinkage. In Phase IIa, it clearly shows chromatin condensation and margination. In Phase IIb, it visualizes nuclear fragmentation and the precise structure of apoptotic bodies [14].
Fluorescence/Confocal Microscopy
- Methodology: Cells are stained with fluorescent DNA-binding dyes such as Hoechst 33342, DAPI (4',6-diamidino-2-phenylindole), or Acridine Orange (AO). These dyes bind to DNA and allow for the visualization of nuclear morphology through fluorescence or confocal microscopy [14].
- Detectable Features: The intensity and distribution pattern of the fluorescence signal are used to indirectly assess nuclear integrity and chromatin condensation. This technique is mainly suitable for observing the nuclear disintegration characteristic of Phase IIb apoptosis [14].
Imaging Flow Cytometry (IFC)
- Methodology: IFC merges the high-throughput, statistical power of conventional flow cytometry with the morphological insight of digital microscopy [49] [50]. Cells in suspension are hydrodynamically focused and passed in single file through one or more laser beams. As each cell passes, it is imaged at high speed and resolution across multiple channels [51] [49].
- Detectable Features: IFC can quantify morphological and fluorescence features across thousands of cells. It can detect cell shrinkage (Phase I) and, with appropriate nuclear stains, can identify chromatin condensation and nuclear fragmentation (Phases IIa and IIb). A key advantage is its ability to provide spatial information, such as protein localization within subcellular compartments, which is lost in conventional flow cytometry [49] [50].
Intravital Microscopy (IVM)
- Methodology: Advanced in vivo imaging techniques, such as two-photon intravital microscopy (2P-IVM), are used to study cell death within the physiological environment of a living animal [52] [53]. This generates 4D time-lapse data (x, y, z, time) of cellular dynamics in organs like the spleen or lymph nodes.
- Detectable Features: IVM can track the entire progression of apoptosis in real-time, including membrane blebbing and the formation of apoptotic bodies in vivo [52]. However, its throughput is limited compared to in vitro techniques [52].
Automated Live-Cell Imaging & Deep Learning
- Methodology: This approach involves continuous live-cell imaging to generate time-lapse data. Advanced computational tools, particularly deep learning models, are then trained to automatically detect and quantify apoptotic events based on morphological hallmarks [53].
- Detectable Features: Systems like ADeS (Apoptosis Detection System) can identify the location and duration of multiple apoptotic events in full microscopy time-lapses by recognizing key morphological changes across all phases, from initial shrinkage to apoptotic body formation [53]. These tools can achieve classification accuracy above 98% and surpass human performance in detection tasks [53].
Comparative Analysis of Techniques
The following tables summarize the key characteristics, advantages, and limitations of each morphological detection technique, providing a clear guide for selection.
Table 1: Technical Specifications and Phase Detection Capabilities
| Technique | Key Readouts | Primary Apoptotic Phase Detected | Throughput | Resolution |
|---|---|---|---|---|
| Light Microscopy | Cell rounding, apoptotic bodies [14] | Phase IIb [14] | Low to Medium | Low (~200 nm) |
| Electron Microscopy | Ultrastructural details, vacuolation, chromatin margination [14] | Phases I, IIa, IIb [14] | Very Low | Very High (~1 nm) |
| Fluorescence Microscopy | Nuclear condensation, fragmentation [14] | Phase IIb [14] | Low to Medium | High (~180 nm) |
| Imaging Flow Cytometry | Cell size, nuclear morphology, protein localization [51] [49] | Phases I, IIa, IIb (population-based) | High (up to 5,000 cells/sec) [51] | Medium (20x/40x objective) [49] |
| Intravital Microscopy | Spatiotemporal dynamics of cell death in vivo [52] | Phases I, IIa, IIb (in context) | Very Low | Medium-High |
| Deep Learning (ADeS) | Automated detection of full apoptotic sequence [53] | Phases I, IIa, IIb (automated) | Medium (post-acquisition) | Dependent on source imaging |
Table 2: Advantages, Limitations, and Suitability for Research Goals
| Technique | Key Advantages | Key Limitations / Potential for False Positives | Best Suited For |
|---|---|---|---|
| Light Microscopy | Simple, convenient, intuitive, storable specimens [14] | Low resolution; cannot detect early phases; small areas of apoptosis easily missed [14] | Initial, low-cost screening for late-stage apoptosis in cell cultures or histology. |
| Electron Microscopy | Reveals definitive, high-resolution ultrastructure; gold standard for morphology [14] | Cannot rule out apoptosis if classic features are absent; requires complementary methods [14] | Detailed ultrastructural analysis to confirm ambiguous results from other methods. |
| Fluorescence Microscopy | Direct visualization of nuclear changes; compatible with live-cell imaging [14] | Small areas of apoptosis easily missed; primarily limited to late-stage nuclear changes [14] | Assessing nuclear morphology and chromatin condensation in fixed or live cells. |
| Imaging Flow Cytometry | High-throughput, quantitative, provides spatial context & statistical power [51] [49] | Higher instrument cost; more complex data analysis than conventional flow cytometry [54] | High-throughput screening, rare event analysis, and phenotyping in heterogeneous populations. |
| Intravital Microscopy | Studies apoptosis in physiological context; reveals cell-cell interactions [52] | Very low throughput; technically challenging; limited availability of open datasets [52] | Understanding the spatial-temporal regulation of apoptosis in living organisms. |
| Deep Learning (ADeS) | High accuracy (>98%); automated; surpasses human performance; label-free potential [53] | "Data-hungry"; requires large, curated datasets for training [53] | Unbiased, high-content analysis of large-scale live-cell imaging datasets. |
Experimental Protocols for Key Techniques
To ensure reproducible results, following standardized protocols is essential. Below are detailed methodologies for two commonly used and highly powerful techniques.
Protocol: Apoptosis Detection via Imaging Flow Cytometry
This protocol is ideal for quantifying morphological features of apoptosis across a large cell population [49] [50].
Cell Preparation and Staining:
- Harvest cells and wash with PBS.
- Resuspend cell pellet in Annexin V binding buffer. A widely used consumable for this purpose is the Annexin V-FITC Apoptosis Detection Kit, where Annexin V-FITC binds to phosphatidylserine exposed on the outer membrane of apoptotic cells [54].
- Add Annexin V-FITC and incubate for 15 minutes in the dark.
- Add a viability dye, such as Propidium Iodide (PI), just before analysis to distinguish late apoptotic and necrotic cells [54].
- For nuclear morphology, add a DNA stain like Hoechst 33342.
Data Acquisition on IFC:
- Calibrate the IFC instrument according to manufacturer instructions.
- Set up acquisition templates to capture brightfield, darkfield, and fluorescence channels for all dyes used.
- Acquire data for a statistically significant number of cells (e.g., 10,000 events per sample).
Data Analysis:
- Use dedicated software to gate on single, focused cells using brightfield gradient RMS.
- Identify apoptotic populations by creating a scatter plot of Annexin V-FITC intensity vs. PI intensity.
- Apply morphological feature analysis (e.g., nuclear spot count, nuclear texture, cell size) to the gated populations to quantify features like chromatin condensation and cell shrinkage.
Protocol: Automated Detection with ADeS Deep Learning Model
This protocol outlines the workflow for using the ADeS deep learning system to detect apoptosis in live-cell imaging data [53].
Dataset Curation:
- Acquire time-lapse microscopy videos (either in vitro or in vivo via intravital microscopy) depicting cells undergoing apoptosis.
- Manually annotate the videos, marking the spatial location and temporal duration of apoptotic events based on morphological hallmarks (cell shrinkage, membrane blebbing, apoptotic body formation).
Model Training:
- Pre-process the image data, which may include cropping, normalization, and sequence generation.
- Split the annotated data into training, validation, and test sets.
- Train the ADeS transformer-based deep learning architecture on the training set. ADeS utilizes activity recognition principles to classify sequences of cellular activity.
Apoptosis Detection and Quantification:
- Input new, unlabeled microscopy time-lapses into the trained ADeS model.
- The model outputs the precise location (x, y coordinates) and duration (start and end frame) of all apoptotic events within the video.
- Use these outputs to quantify metrics such as apoptosis rate, timing, and spatial distribution within the tissue or culture.
Visualization of Workflows and Relationships
To clarify the decision-making process and technical relationships, the following diagrams illustrate the experimental selection workflow and the core principle of Imaging Flow Cytometry.
Diagram 1: A flowchart to guide the selection of an appropriate morphological detection technique based on key research questions and priorities.
Diagram 2: Core components and workflow of an Imaging Flow Cytometry system, illustrating how cells are imaged at high speed to generate quantitative morphological data.
The Scientist's Toolkit: Key Reagent Solutions
Successful morphological analysis relies on a suite of essential reagents and tools. The following table details key solutions used in the field.
Table 3: Essential Research Reagents and Materials for Morphological Apoptosis Detection
| Reagent/Material | Function/Application | Example Use Case |
|---|---|---|
| Annexin V-FITC/PI Kit | Flags early (Annexin V+/PI-) and late (Annexin V+/PI+) apoptotic cells by detecting phosphatidylserine exposure and membrane integrity [54]. | Standard assay for flow cytometry and imaging flow cytometry to quantify apoptotic populations. |
| Hoechst 33342 / DAPI | Cell-permeant (Hoechst) and cell-impermeant (DAPI) DNA stains for visualizing nuclear morphology and chromatin condensation [14]. | Fluorescence microscopy to identify pyknosis and karyorrhexis; counterstain for cell counting. |
| Caspase Activity Probes | Fluorescently-labeled inhibitors or substrates that covalently bind to active caspase enzymes, key executioners of apoptosis. | Validating the engagement of apoptotic pathways in conjunction with morphological changes. |
| Mitochondrial Potential Dyes | Cationic dyes (e.g., JC-1, TMRM) that accumulate in active mitochondria; a shift from red to green fluorescence indicates loss of membrane potential, an early apoptotic event [14]. | Detecting early Phase I apoptosis via the mitochondrial pathway using fluorescence microscopy or flow cytometry. |
| Imaging Flow Cytometer | Instrument that combines high-throughput flow analysis with high-resolution cellular imaging [49] [50]. | Acquiring quantitative morphological data from thousands of cells for statistical analysis of apoptosis. |
| ADeS Software | A deep learning-based algorithm for automated detection of apoptosis in live-cell imaging data [53]. | Unbiased, high-throughput analysis of the location and duration of apoptosis in microscopy time-lapses. |
Correlating Morphological Findings with Biochemical Assays (e.g., Caspase Activation)
Apoptosis, or programmed cell death, is a fundamental biological process crucial for tissue development, homeostasis, and the elimination of damaged or harmful cells. Its dysregulation is implicated in a spectrum of diseases, including cancer, neurodegenerative disorders, and autoimmune conditions [14]. Research into apoptotic mechanisms is thus a cornerstone of modern biomedical science, particularly in drug discovery and development.
A comprehensive understanding of apoptosis requires the integration of multiple analytical perspectives. The process is characterized by a tightly coupled sequence of morphological changes and biochemical events [1]. Relying on a single detection method can yield an incomplete or misleading picture. This guide provides an in-depth technical framework for the systematic correlation of the classic morphological features of apoptosis (Phases I, IIa, and IIb) with key biochemical assays, with a particular focus on caspase activation. This integrated approach is essential for confirming the occurrence of apoptosis, distinguishing it from other modes of cell death like necrosis, and precisely delineating the stage-specific progression of cell demise within the context of a broader research thesis on apoptotic morphology [14].
Morphological Hallmarks of Apoptosis Phases I, IIa, and IIb
The morphological progression of apoptosis is typically divided into distinct phases, each with characteristic features that can be visualized using various microscopic techniques [14].
Table 1: Morphological Characteristics of Apoptotic Phases [14]
| Apoptotic Phase | Key Morphological Features | Primary Detection Methods |
|---|---|---|
| Phase I (Early) | Cell shrinkage, condensed cytoplasm, loss of microvilli, surface blebbing, vacuolation (cavitation). | Transmission Electron Microscopy (TEM) |
| Phase IIa (Intermediate) | Nuclear chromatin condensation (pyknosis), chromatin margination (assembly on inner nuclear membrane). | Fluorescence/Confocal Microscopy (Hoechst, DAPI), TEM |
| Phase IIb (Late) | Nuclear fragmentation (karyorrhexis), formation of membrane-bound apoptotic bodies. | Light Microscopy (HE, Giemsa staining), TEM, Fluorescence Microscopy |
Figure 1: The Morphological Progression of Apoptosis. This workflow illustrates the sequential stages of apoptosis, from the initial trigger to the final clearance of cellular debris, highlighting key morphological events at each phase [14].
Key Biochemical Assays for Apoptosis Detection
Biochemical events run in parallel to morphological changes. Key assays target specific molecular hallmarks of apoptosis, such as caspase activation, phosphatidylserine externalization, and DNA fragmentation [14] [1].
Table 2: Key Biochemical Assays for Apoptosis Detection [14]
| Assay | Principle | Target Process | Apoptotic Stage | Key Advantages | Key Limitations |
|---|---|---|---|---|---|
| Caspase Activity Assay | Measures cleavage of fluorogenic or chromogenic substrates by active caspase enzymes. | Caspase activation (Intrinsic/Extrinsic Pathways) | Early to Mid | Quantitative, high-throughput capability, pathway-specific. | Does not confirm cell death commitment; early event. |
| Annexin V Staining | Binds to phosphatidylserine (PS) translocated to the outer leaflet of the plasma membrane. | Loss of membrane asymmetry | Early (before membrane integrity loss) | Distinguishes early apoptosis (Annexin V+/PI-) from late apoptosis/necrosis (Annexin V+/PI+). | Requires flow cytometry or fluorescence microscopy; can be non-specific under certain conditions. |
| TUNEL Assay | Terminal deoxynucleotidyl transferase (TdT) labels 3'-OH ends of fragmented DNA. | DNA fragmentation | Mid to Late | Highly specific for apoptosis in standard conditions; allows in-situ detection. | Can yield false positives in necrotic cells or with DNA damage; not suitable for early apoptosis. |
| DNA Gel Electrophoresis | Detects internucleosomal DNA cleavage into a "ladder" pattern of ~180-200 bp fragments. | DNA fragmentation | Mid to Late | Simple, qualitatively accurate. | Low sensitivity; requires large number of apoptotic cells; cannot localize apoptotic cells. |
| Mitochondrial Membrane Potential (ΔΨm) Assay | Uses fluorescent dyes (e.g., JC-1) that accumulate in mitochondria in a potential-dependent manner. | Mitochondrial outer membrane permeabilization (MOMP) | Early (Intrinsic Pathway) | Early marker of intrinsic apoptosis; can be performed with flow cytometry. | Sensitive to cell health and assay conditions (e.g., pH). |
Figure 2: Core Apoptotic Signaling Pathways. This diagram outlines the major signaling cascades in apoptosis, culminating in the activation of executioner caspases and the manifestation of characteristic biochemical and morphological hallmarks [14] [1].
Integrated Experimental Protocols for Correlation
This section provides detailed methodologies for experiments designed to directly correlate morphological observations with biochemical data.
Protocol: Correlating Phase IIa/IIb Morphology with Caspase-3 Activation
Objective: To confirm that cells displaying chromatin condensation and nuclear fragmentation are undergoing caspase-dependent apoptosis.
Materials:
- Cells of interest treated with apoptotic inducer and controls.
- Caspase-3 fluorogenic substrate (e.g., DEVD-AFC or DEVD-AMC).
- Fluorescent DNA stain (e.g., Hoechst 33342 or DAPI).
- Cell culture facilities, fluorescence microplate reader, fluorescence/confocal microscope.
Method:
- Treat Cells: Seed cells in multiple plates/wells. Apply apoptotic inducer for a determined time course (e.g., 0, 2, 6, 12, 24 hours).
- Harvest Cells: For each time point, collect both adherent and floating cells.
- Caspase-3 Activity Assay (Biochemical):
- Lyse a portion of the cell pellet.
- Incubate cell lysate with caspase-3 substrate in assay buffer.
- Measure fluorescence (e.g., AFC: Ex~400 nm, Em~505 nm) in a microplate reader at timed intervals.
- Normalize fluorescence values to total protein concentration.
- Nuclear Morphology Assessment (Morphological):
- For parallel samples, seed cells on chambered slides.
- At corresponding time points, stain live cells with Hoechst 33342 (1-2 µg/mL) for 20 minutes.
- Wash and immediately image using a fluorescence microscope with a DAPI filter set.
- Score at least 200 cells per condition for normal nuclei, condensed chromatin (Phase IIa), and fragmented nuclei (Phase IIb).
Correlation Analysis:
- Plot the time course of caspase-3 activity (relative fluorescence units/µg protein) alongside the percentage of cells exhibiting Phase IIa and IIb morphology.
- A strong positive correlation, where rising caspase activity precedes or coincides with an increase in nuclear condensation/fragmentation, robustly confirms apoptotic execution.
Protocol: Annexin V/Propidium Iodide (PI) Staining Coupled with Morphological Analysis
Objective: To distinguish early apoptotic (morphologically intact) from late apoptotic/necrotic cells.
Materials:
- Annexin V-FITC conjugate (or other fluorophore).
- Propidium Iodide (PI) stock solution.
- Binding buffer (typically 10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl₂, pH 7.4).
- Flow cytometer and fluorescence microscope.
Method:
- Induce and Harvest: Treat and harvest cells gently to avoid mechanical damage.
- Stain Cells: Resuspend cell pellet in binding buffer containing Annexin V-FITC and PI as per manufacturer's instructions. Incubate for 10-15 minutes in the dark.
- Analyze by Flow Cytometry (Biochemical/Phenotypic):
- Analyze cells on a flow cytometer.
- Identify populations: Annexin V-/PI- (viable), Annexin V+/PI- (early apoptotic), Annexin V+/PI+ (late apoptotic/necrotic).
- Correlative Microscopy (Morphological):
- After staining, place a drop of cell suspension on a slide and coverslip.
- Immediately image using a fluorescence microscope with FITC and TRITC/Rhodamine filter sets.
- Correlate: Cells that are Annexin V+/PI- (early apoptotic) should appear morphologically intact, possibly with early signs of shrinkage or blebbing (Phase I). Cells that are Annexin V+/PI+ should show advanced apoptotic morphology like nuclear condensation (Phase IIa) or fragmentation (Phase IIb), or necrotic characteristics.
The Scientist's Toolkit: Essential Reagents and Kits
The apoptosis assay market offers a wide range of standardized, commercially available kits that facilitate robust and reproducible detection. The market is projected to grow significantly, driven by rising R&D in chronic diseases and drug discovery [54] [55].
Table 3: Essential Research Reagent Solutions for Apoptosis Detection [54] [56] [55]
| Product Category | Example Products/Kits | Primary Function | Key Characteristics |
|---|---|---|---|
| Caspase Activity Assay Kits | Caspase-Glo 3/7 Assay; Fluorometric Caspase-3 Assay Kits | Measure proteolytic activity of specific caspases using luminescent or fluorogenic substrates. | Highly sensitive, quantitative, amenable to high-throughput screening. |
| Annexin V Staining Kits | Annexin V-FITC Apoptosis Detection Kits (e.g., from Merck, Thermo Fisher) | Detect phosphatidylserine externalization on the outer plasma membrane. | Often include PI for viability staining; compatible with flow cytometry and microscopy. |
| TUNEL Assay Kits | In Situ Cell Death Detection Kits (e.g., from Roche) | Label DNA strand breaks for in-situ detection of apoptotic nuclei. | Allows visualization and quantification of apoptosis in cell cultures and tissue sections. |
| Mitochondrial Potential Assays | JC-1 Assay Kits; TMRE Staining Kits | Detect loss of mitochondrial membrane potential (ΔΨm). | JC-1 exhibits potential-dependent emission shift (redgreen); early apoptosis indicator. |
| Multiplex Apoptosis Assay Kits | Multiplex Assays combining caspase activity with other markers (e.g., CellEvent Caspase-3/7 Green) | Enable simultaneous detection of multiple apoptotic events in a single sample. | Provides more comprehensive data, saves sample, reduces assay variability. |
Data Interpretation and Correlation Strategy
Successfully integrating morphological and biochemical data requires a strategic approach to interpretation.
- Temporal Correlation is Key: Biochemical events often precede visible morphological changes. For instance, caspase activation and phosphatidylserine externalization are early events that occur before overt nuclear fragmentation [14] [1]. A time-course experiment is therefore far more informative than a single endpoint measurement.
- Quantitative Over Qualitative: Whenever possible, quantify both morphological and biochemical data. Instead of "more condensed nuclei," report "a 40% increase in cells with pyknotic nuclei." This allows for statistical correlation with biochemical readouts like caspase activity units.
- Use Controls Rigorously: Include appropriate controls (e.g., untreated cells, cells treated with a known apoptotic inducer, and caspase inhibitor-treated cells) to validate the specificity of your assays and interpretations.
- Address Discrepancies: If morphology suggests apoptosis but biochemical assays are negative (or vice versa), investigate potential pitfalls. For example, the TUNEL assay can give false positives in necrotic cells, and caspase-independent apoptosis pathways exist [14]. Using multiple, orthogonal assays strengthens the overall conclusion.
Figure 3: Integrated Data Analysis Workflow. A logical flow for designing and executing experiments that correlate biochemical and morphological data to draw robust conclusions about the apoptotic status of cells.
Overcoming Challenges in Morphological Apoptosis Detection
Accurate distinction between apoptosis, necrosis, and autophagic cell death is a fundamental requirement in biomedical research, with critical implications for understanding disease mechanisms, developing therapeutic interventions, and evaluating drug efficacy. Within the context of broader thesis research on morphological features of apoptosis phases I, IIa, and IIb, this technical guide provides a comprehensive framework for differentiating these primary cell death modalities. The morphological hallmarks of apoptotic phases establish a critical foundation for this discrimination: Phase I involves cell shrinkage and cytoplasmic condensation; Phase IIa features nuclear chromatin condensation and margination; and Phase IIb demonstrates nuclear fragmentation and apoptotic body formation [14] [57]. These precise morphological changes contrast sharply with the cellular swelling and membrane rupture characteristic of necrosis, and the vacuolization typical of autophagic cell death [17]. This guide synthesizes current methodologies, molecular mechanisms, and detection strategies to empower researchers in making accurate distinctions between these pathways, thereby enhancing experimental validity and biological interpretation.
Morphological Characteristics: A Comparative Analysis
The most fundamental approach to distinguishing cell death types begins with detailed morphological assessment across multiple imaging modalities. Different cell death pathways create distinctive structural alterations in cells, providing initial diagnostic clues that should be confirmed with biochemical assays.
Table 1: Comparative Morphological Features of Major Cell Death Types
| Feature | Apoptosis | Necrosis/Necroptosis | Autophagic Cell Death |
|---|---|---|---|
| Cell Size | Shrinkage (pylknosis) [14] | Swelling (oncosis) [17] [58] | Variable (may be reduced) |
| Nucleus | Chromatin condensation (Phase IIa), fragmentation (Phase IIb) [14] [17] | Karyolysis (clumping, then dissolution) [17] | Less obvious pyknosis than apoptosis [17] |
| Plasma Membrane | Blebbing, integrity maintained [20] [57] | Rapid rupture, loss of integrity [20] [17] | Integrity largely maintained |
| Membrane Asymmetry | Phosphatidylserine (PS) externalization [57] | No controlled PS exposure | Not a defining feature |
| Cytoplasm | Condensation, organelle packaging [57] | Dilatation of organelles, content leakage [20] | Abundant autophagic vacuoles [17] |
| Key Inclusions | Apoptotic bodies [14] [57] | None | Double-membrane autophagosomes [6] |
| Inflammatory Response | None (clean removal) [57] | Significant (DAMP release) [17] | Generally non-inflammatory |
| Elimination | Phagocytosis by neighboring cells [14] | Cell lysis in situ [17] | Lysosomal degradation (autolysosomes) [6] |
Advanced label-free imaging techniques like Quantitative Phase Imaging (QPI) and Full-Field Optical Coherence Tomography (FF-OCT) now enable researchers to monitor these morphological dynamics in real-time without fixation or staining artifacts. QPI detects apoptosis through characteristic cell shrinkage and the appearance of a sharp nuclear boundary, while necrosis presents with cell swelling and a gradual loss of intracellular density [59] [58]. FF-OCT provides ultra-high-resolution tomography, revealing subcellular features such as the echinoid spine formation and membrane blebbing in apoptosis versus rapid membrane rupture in necrosis [20].
Molecular Mechanisms and Signaling Pathways
Beyond morphology, each cell death pathway is defined by distinct molecular machinery and signaling cascades. Understanding these mechanisms provides the foundation for developing specific biochemical assays and molecular markers.
Apoptosis Signaling Pathways
Apoptosis proceeds through two well-defined molecular pathways that converge on a common execution phase:
Extrinsic (Death Receptor) Pathway: Initiated by extracellular death ligands (e.g., FasL, TNF-α) binding to cell surface receptors, leading to the formation of the Death-Inducing Signaling Complex (DISC). This complex activates initiator caspase-8 and caspase-10, which then activate executioner caspases-3, -6, and -7 [6] [60].
Intrinsic (Mitochondrial) Pathway: Triggered by intracellular stress signals (DNA damage, oxidative stress), leading to mitochondrial outer membrane permeabilization (MOMP) controlled by the Bcl-2 protein family balance. This results in cytochrome c release, apoptosome formation with Apaf-1, and activation of caspase-9, which then activates executioner caspases [61] [60].
Both pathways ultimately activate executioner caspases-3 and -7, which cleave key cellular substrates including PARP, leading to the characteristic morphological changes of apoptosis [17] [57].
Diagram 1: Molecular signaling pathways in apoptosis, necroptosis, and autophagic cell death. Execution phases highlighted in red indicate irreversible commitment to cell death.
Necroptosis and Autophagic Cell Death Pathways
Necroptosis: This regulated form of necrosis shares initiation triggers with apoptosis (e.g., TNF receptor activation) but proceeds through a distinct molecular pathway when caspase-8 is inhibited. Key mediators include receptor-interacting protein kinases RIPK1 and RIPK3, which phosphorylate the mixed lineage kinase domain-like protein (MLKL). Phosphorylated MLKL oligomerizes and translocates to the plasma membrane, causing membrane rupture and release of damage-associated molecular patterns (DAMPs) that trigger inflammation [17] [62].
Autophagic Cell Death: Characterized by the accumulation of autophagic vacuoles, this process involves the ULK1 complex activation, often through mTOR inhibition under stress conditions. This triggers phagophore formation, which elongates through two ubiquitin-like conjugation systems (ATG5-ATG12 and LC3-I to LC3-II conversion) to form double-membrane autophagosomes. Fusion with lysosomes creates autolysosomes where cellular components are degraded [61] [6]. While autophagy typically promotes survival, its hyperactivation can lead to cell death through excessive self-digestion.
Detection Methodologies and Experimental Protocols
Accurate discrimination of cell death modalities requires integrating multiple complementary techniques. The following section details key experimental approaches for identifying and quantifying different cell death pathways.
Imaging-Based Detection Protocols
Label-Free Quantitative Phase Imaging (QPI)
- Principle: Measures phase shifts of light passing through cells to quantify biomass distribution and density without staining [59] [58].
- Protocol Setup: Use a diffraction phase microscopy system with a 532-nm laser source and 20× objective (NA 0.5). Maintain cells at 37°C with 5% CO₂ during time-lapse imaging [59].
- Key Parameters: Track cell area, circularity, optical volume (dry mass), and nuclear edge score. Apoptotic cells show decreasing area and increasing nuclear edge sharpness, while necrotic cells demonstrate swelling followed by sudden collapse [59].
- Data Analysis: Apply sigmoid function fitting to parameter dynamics. Use machine learning classifiers (Support Vector Machines) to distinguish death subtypes with >75% accuracy based on morphological dynamics [59] [58].
Full-Field Optical Coherence Tomography (FF-OCT)
- Principle: Interferometric technique providing high-resolution 3D tomography of cellular structures using broadband light sources [20].
- System Configuration: Custom-built time-domain FF-OCT with halogen light source (650 nm center wavelength), Linnik interferometer with identical 40× water-immersion objectives (NA 0.8), and CCD camera detection [20].
- Processing: Generate en face tomographic images through phase-shifting interference and 3D surface topography via maximum intensity z-position mapping [20].
- Application: Visualize apoptotic membrane blebbing and filopodia reorganization versus necrotic membrane rupture and adhesion loss [20].
Biochemical and Molecular Detection Assays
Table 2: Key Biochemical Assays for Cell Death Discrimination
| Assay Type | Target Process | Apoptosis Readout | Necrosis/Necroptosis Readout | Autophagy Readout |
|---|---|---|---|---|
| Western Blot | Protein cleavage/activation [57] | Cleaved caspases-3, -7, -9; Cleaved PARP [57] | Phospho-RIPK1, RIPK3, MLKL [62] | LC3-I to LC3-II conversion; p62 degradation [6] |
| Fluorescence Microscopy | Membrane integrity/ enzyme activity [59] | Caspase-3/7 positive, PI negative (early) [59] | Caspase-3/7 negative, PI positive [59] | LC3 puncta formation; Cyto-ID staining |
| Flow Cytometry | Multiparameter analysis | Annexin V+/PI- (early), Annexin V+/PI+ (late) [14] | Annexin V+/PI+ (primary positive) | LC3 flux analysis with bafilomycin |
| DNA Fragmentation | Nuclear degradation | TUNEL positive (DNA ladder) [14] | Random DNA degradation (smear) | Not applicable |
| Mitochondrial Function | Membrane potential | JC-1 green fluorescence (ΔΨm loss) [14] | Not specific | Not primary marker |
Western Blot Protocol for Apoptosis Detection
- Sample Preparation: Lyse cells in RIPA buffer with protease and phosphatase inhibitors. Quantify protein concentration to ensure equal loading [57].
- Electrophoresis and Transfer: Separate 20-50 μg protein extract on 4-20% gradient SDS-PAGE gels. Transfer to PVDF membranes using standard protocols [57].
- Antibody Probing: Block with 5% non-fat milk, then incubate with primary antibodies (cleaved caspase-3, cleaved PARP, β-actin loading control) overnight at 4°C. Use HRP-conjugated secondary antibodies and chemiluminescent detection [57].
- Analysis: Normalize band intensity to loading controls. Calculate cleaved-to-total protein ratios (e.g., cleaved caspase-3/total caspase-3) to quantify activation [57].
Multiparameter Fluorescence Staining Protocol
- Cell Preparation: Seed cells in chambered coverslips. Apply death inducers with/without inhibitors (e.g., z-VAD-FMK for pan-caspase inhibition) [58].
- Staining: Load with CellEvent Caspase-3/7 Green reagent (2 μM) and propidium iodide (1 μg/mL). Include Hoechst 33342 for nuclear morphology [59] [58].
- Image Acquisition: Use multimodal microscopy with environmental control (37°C, 5% CO₂). Acquire images every 30-60 minutes for time-lapse tracking [58].
- Classification: Apoptotic cells: Caspase-3/7+, PI- (early) → PI+ (late). Necrotic cells: Caspase-3/7-, PI+ [59].
Research Reagent Solutions
Selecting appropriate reagents is crucial for specific and accurate cell death detection. The following toolkit summarizes essential materials and their applications.
Table 3: Essential Research Reagents for Cell Death Detection
| Reagent Category | Specific Examples | Application & Function | Key Considerations |
|---|---|---|---|
| Inducers | Doxorubicin (5 μM) [20]; Staurosporine (0.5 μM) [58]; Ethanol (99%) [20]; Hydrogen Peroxide (600-700 μM) [59] | Trigger specific death pathways; Doxorubicin: intrinsic apoptosis; Ethanol: necrosis | Concentration-dependent effects; Cell type-specific responses |
| Inhibitors | z-VAD-FMK (10-20 μM) [58]; Necrostatin-1 (RIPK1 inhibitor) | Pathway inhibition to confirm mechanism; z-VAD-FMK: pan-caspase inhibitor | Verify specificity with multiple assays; Potential off-target effects |
| Antibodies | Cleaved caspase-3; Cleaved PARP; Phospho-MLKL; LC3B [57] | Western blot, immunofluorescence; Detect key pathway-specific markers | Validate antibodies for specific applications; Check species reactivity |
| Viability Stains | Propidium Iodide; Ethidium Homodimer III [59] | Membrane integrity assessment; PI exclusion in viable cells | Distinguish late apoptosis vs. necrosis; Use with caspase markers |
| Activity Probes | CellEvent Caspase-3/7 Green [59]; Annexin V conjugates [57] | Detect caspase activation; PS externalization (early apoptosis) | Combine with viability stains for staging; Optimize timing for detection |
| Autophagy Probes | Cyto-ID; LC3-GFP constructs; Lysotracker | Autophagosome visualization; Lysosomal tracking | Monitor flux with bafilomycin A1; Distinguish from apoptosis |
The accurate discrimination between apoptosis, necrosis, and autophagic cell death requires a multifaceted approach integrating morphological assessment, molecular analysis, and functional assays. Researchers should prioritize orthogonal verification using at least two complementary methods, such as combining QPI's dynamic morphological tracking with Western blot analysis of pathway-specific markers. The expanding understanding of regulated necrosis forms like necroptosis, plus the nuanced relationship between autophagy and cell death, underscores the importance of rigorous experimental design. As research continues to reveal complex cross-talk between cell death pathways, the methodologies outlined in this guide provide a foundation for precise differentiation, enabling more accurate biological interpretation and enhancing the validity of therapeutic screening in drug development pipelines.
Addressing False Positives and Negatives in Morphological Analysis
The accurate assessment of programmed cell death (PCD) is fundamental to biomedical research, particularly in oncology and drug development. Among PCD modalities, apoptosis remains the most extensively characterized, serving as a crucial indicator for treatment efficacy and toxicology studies. Morphological analysis provides the definitive "gold standard" for identifying apoptotic cells and distinguishing between different modes of cell death [63] [64]. However, the accurate morphological identification of apoptosis, particularly during its distinct phases (I, IIa, and IIb), is frequently compromised by false positives and negatives, potentially leading to erroneous conclusions in both basic research and clinical applications.
The inherent challenges stem from several factors: the transient nature of morphological changes, the coexistence of multiple cell death pathways, technical artifacts introduced during sample preparation, and the subjective interpretation of cellular features. This technical guide examines the principal sources of error in the morphological analysis of apoptosis and provides researchers with robust methodological frameworks to enhance the reliability and reproducibility of their findings within the broader context of apoptosis phase research.
Morphological Hallmarks of Apoptosis and Its Distinct Phases
Core Morphological Criteria
Apoptosis is characterized by a conserved sequence of morphological alterations that distinguish it from other forms of cell death, such as necrosis. The process typically initiates with cell shrinkage and chromatin condensation (pyknosis), progresses to nuclear fragmentation (karyorrhexis), and culminates in the formation of membrane-bound apoptotic bodies that are rapidly phagocytosed by neighboring cells without provoking an inflammatory response [65] [64]. These morphological features result from the coordinated activation of biochemical executioners, primarily caspases, which systematically dismantle cellular structures [13].
The key distinguishing feature between apoptosis and necrosis lies in membrane integrity. Throughout most of the apoptotic process, the plasma membrane remains intact, effectively containing intracellular contents and preventing inflammation. In contrast, necrotic cells undergo immediate loss of membrane integrity, leading to the release of cellular components and subsequent inflammatory reactions [65] [17].
Phase-Specific Morphological Transitions
Advanced morphological analysis recognizes distinct phases within the apoptotic continuum, each with characteristic features that can be precisely identified.
Table 1: Morphological Characteristics Across Apoptosis Phases
| Phase | Key Morphological Features | Cellular & Nuclear Changes | Duration | Primary Detection Methods |
|---|---|---|---|---|
| Phase I (Early) | Cell shrinkage, chromatin condensation (pyknosis), cytoplasmic condensation | Preservation of organelle integrity, ribosome detachment from endoplasmic reticulum | Variable (minutes to hours) | Electron microscopy, vital dye exclusion, Annexin V staining |
| Phase IIa (Mid) | Nuclear fragmentation (karyorrhexis), pronounced membrane blebbing | Activation of executioner caspases, cleavage of structural proteins | ~2 hours in vitro [64] | Light microscopy, caspase activation assays, TUNEL assay |
| Phase IIb (Late) | Formation of apoptotic bodies, phagocytosis by adjacent cells | Containment of nuclear fragments and organelles within intact membranes | 12-24 hours in vivo [64] | Light and electron microscopy, TUNEL assay on apoptotic bodies |
The entire apoptotic process, from initiation to clearance, is estimated to last approximately 12-24 hours in vivo, though in vitro the visible morphologic changes may complete in less than two hours [64]. This temporal dynamic is crucial for experimental design, as single timepoint analyses may miss the transient morphological window.
Technical Artifacts and Methodological Pitfalls
Sample preparation introduces significant artifacts that can be misinterpreted as apoptotic morphology. Chemical fixation can induce cell shrinkage and pyknosis resembling early apoptosis, while mechanical stress from tissue processing may cause membrane disruptions indistinguishable from late-stage apoptotic bodies [63]. The TUNEL assay, while widely used for detecting DNA fragmentation, is particularly prone to false positives from extensive DNA degradation in necrotic cells and false negatives in early apoptosis where DNA cleavage may be incomplete [64]. Furthermore, active RNA synthesis in viable cells and variations in tissue fixation and protease digestion during TUNEL procedures can generate non-specific staining [64].
The apoptosis-necrosis continuum represents another significant challenge, where the same initial insult can trigger different death pathways depending on intensity, duration, and cellular ATP levels. At low doses of injurious stimuli (heat, radiation, hypoxia), cells typically undergo apoptosis, while the same stimuli at higher doses result in necrosis [65]. Cells may also display hybrid morphological features, especially under pathological conditions like ischemia, where the lack of ATP can abort the apoptotic program before full morphological manifestation [64].
Limitations of Single-Parameter Assessment
Relying on a single morphological or biochemical marker inevitably increases vulnerability to misclassification. For instance, DNA fragmentation detected by TUNEL occurs in both apoptosis and necrosis, though the pattern differs—organized apoptotic bodies versus disorganized cellular debris [66]. Similarly, phosphatidylserine externalization, detected by Annexin V binding, can occur in non-apoptotic cells with membrane perturbations [66] [67]. Caspase activation, while central to apoptosis, may not always progress to full execution in the presence of endogenous inhibitors, resulting in incomplete morphological development [13].
The following diagram illustrates the decision pathway for distinguishing true apoptosis from common false positives:
Integrated Methodological Approaches for Accurate Detection
Multiparameter Analytical Framework
Overcoming the limitations of single-parameter assessment requires a multiparameter analytical framework that correlates morphological features with biochemical markers across multiple dimensions. Flow and image cytometry provide ideal platforms for this approach, enabling simultaneous quantification of caspase activation, membrane integrity, mitochondrial membrane potential, and DNA fragmentation in individual cells [63]. This correlative strategy is essential because apoptotic markers manifest in variable constellations depending on cell type, death stimulus, and microenvironmental conditions [63].
A robust experimental workflow should incorporate both temporal and spatial considerations, analyzing multiple time points to capture the progression of morphological changes and examining sufficient microscopic fields to account for heterogeneity. The confirmation of apoptosis requires demonstrating a coherent pattern where early biochemical events (caspase activation, phosphatidylserine exposure) precede and culminate in the characteristic morphological endpoint (apoptotic body formation) [63].
Standardized Experimental Protocols
Protocol 1: Multiparametric Flow Cytometry for Apoptosis Quantification
This protocol enables simultaneous assessment of multiple apoptotic parameters, significantly reducing false positives/negatives:
- Cell Preparation: Harvest approximately 1×10^6 cells per condition, using gentle centrifugation (300 × g for 5 minutes) to minimize mechanical damage.
- Phosphatidylserine Exposure & Membrane Integrity: Resuspend cells in Annexin V binding buffer containing:
- Caspase Activation: For intracellular caspase detection, fix and permeabilize cells using commercial fixation/permeabilization buffers, then incubate with anti-cleaved caspase-3/7 antibodies (1:50 dilution) for 30 minutes at 4°C.
- DNA Fragmentation Analysis: Following surface and intracellular staining, fix cells with 4% paraformaldehyde for 15 minutes, permeabilize with 0.1% Triton X-100, and perform TUNEL staining according to manufacturer's protocol.
- Flow Cytometric Analysis: Acquire data using a flow cytometer capable of detecting at least 3 fluorescence parameters. Analyze populations as follows:
Protocol 2: Morphological Validation by Fluorescence Microscopy
This complementary protocol provides essential morphological confirmation:
- Cell Culture: Plate cells on chambered coverslips or glass coverslips to achieve 60-70% confluence at time of analysis.
- Staining: Load cells with:
- Hoechst 33342 (1 μg/mL) for nuclear morphology assessment
- Annexin V-Cy3 (1:100 dilution) in binding buffer
- SYTOX Green (50 nM) for membrane integrity evaluation
- Incubate for 15-20 minutes at 37°C
- Fixation: For permanent preparations, fix with 4% paraformaldehyde for 15 minutes after live-cell imaging.
- Image Acquisition: Capture images using a fluorescence microscope with 40× or 60× objectives. For each condition, analyze at least 10 random fields containing approximately 100-200 cells each.
- Morphological Scoring: Systematically evaluate:
Essential Research Reagent Solutions
Table 2: Key Research Reagents for Accurate Apoptosis Detection
| Reagent/Category | Specific Examples | Primary Function | Considerations & Limitations |
|---|---|---|---|
| Viability Probes | Propidium Iodide, 7-AAD, SYTOX Green | Membrane integrity assessment through exclusion from viable cells | Cannot distinguish apoptotic vs. necrotic membrane rupture; requires immediate analysis [67] |
| Phosphatidylserine Detection | Annexin V conjugates (FITC, PE, Cy3) | Detection of PS externalization as early apoptotic marker | Requires calcium-containing buffer; can yield false positives in necrotic cells [66] [67] |
| Caspase Activity Assays | Fluorogenic substrates (DEVD-AFC), cleaved caspase antibodies | Specific detection of apoptotic executive machinery activation | May not detect caspase-independent apoptosis pathways [13] [64] |
| DNA Fragmentation Assays | TUNEL assay kits, DNA laddering detection | Detection of internucleosomal DNA cleavage | Prone to false positives in necrotic cells; requires careful standardization [66] [64] |
| Nuclear Morphology Stains | Hoechst 33342, DAPI, DRAQ5 | Visualization of chromatin condensation and nuclear fragmentation | Requires fluorescence microscopy; subjective interpretation [63] |
| Mitochondrial Probes | TMRE, JC-1, MitoTracker Red | Assessment of mitochondrial membrane potential (ΔΨm) | Loss of ΔΨm not exclusive to apoptosis; occurs in necrosis [66] |
Experimental Design and Validation Strategies
Controls and Standardization
Implementing appropriate controls is fundamental for validating morphological assessments and minimizing misinterpretation:
- Positive Controls: Treat cells with established apoptosis inducers:
- Negative Controls: Include untreated, healthy cells from the same population.
- Technical Controls: For TUNEL assays, include:
- DNase I-treated samples (positive control for DNA fragmentation)
- Omission of TdT enzyme (negative control for labeling specificity) [64]
- Necrosis Controls: Induce necrosis by:
Standardization across experiments requires careful attention to fixation methods (avoid over-fixation), antibody concentrations (determined by titration), and consistent imaging parameters. Quantitative analysis should incorporate sufficient biological replicates (minimum n=3) and adequate cell numbers (typically 10,000 events for flow cytometry, 200+ cells for microscopy) to ensure statistical robustness [63].
Advanced Technical Approaches
For complex research questions, consider these advanced methodologies:
- Time-Lapse Microscopy: Monitors dynamic morphological changes in individual cells, capturing transient intermediate states that might be missed in endpoint assays [63].
- Electron Microscopy: Provides ultrastructural details for definitive morphological classification, particularly useful for distinguishing ambiguous cases [65] [64].
- High-Content Screening Systems: Combine automated microscopy with multiparameter image analysis to quantitatively assess morphological features across thousands of cells under different experimental conditions [68].
- Caspase Inhibition Experiments: Use pan-caspase inhibitors (e.g., Z-VAD-FMK) to determine caspase dependence of observed cell death morphology [64].
The following workflow integrates these methodologies into a comprehensive strategy for minimizing false interpretations:
The accurate morphological analysis of apoptosis phases demands a methodical, integrated approach that acknowledges and addresses the multiple potential sources of false positives and negatives. By implementing the multiparameter frameworks, standardized protocols, and rigorous validation strategies outlined in this guide, researchers can significantly enhance the reliability of their apoptosis assessments. This methodological rigor is particularly crucial in translational applications, including drug discovery and therapeutic development, where accurate cell death quantification directly impacts decision-making and clinical translation. As cell death research continues to evolve, maintaining morphological analysis as the foundational gold standard—while complementing it with biochemical and molecular techniques—will ensure the continued generation of robust, reproducible data in apoptosis research.
Strategies for Detecting Apoptosis in Small or Rapidly Cleared Cell Populations
The morphological features of apoptosis are classically divided into three phases: Phase I (cell shrinkage, dense cytoplasm), Phase IIa (chromatin condensation and margination), and Phase IIb (nuclear fragmentation and apoptotic body formation) [14]. A significant challenge in cell death research is the reliable detection of apoptotic events when the cell population is small or when apoptotic cells are rapidly cleared. The phagocytosis of apoptotic cells is very effective and rapid; consequently, apoptotic cells are quickly removed without leaving traces, making apoptosis in a small area difficult to recognize through morphology alone [14]. This technical guide outlines sophisticated strategies to overcome these limitations, providing researchers and drug development professionals with robust tools for accurate apoptosis detection in challenging scenarios.
Technical Approaches and Their Applications
The selection of an appropriate detection method must account for the specific apoptotic phase being investigated and the technical constraints related to sample size and cell clearance kinetics. The following table summarizes the core characteristics of advanced detection methods suitable for limited populations.
Table 1: Apoptosis Detection Methods for Small or Rapidly Cleared Populations
| Method | Principle | Target Apoptotic Phase | Key Advantage for Small Populations | Sample Requirement |
|---|---|---|---|---|
| Flow Cytometry (Multiparameter) [69] | Multi-parameter analysis at single-cell level (e.g., Δψm, caspases, Annexin V) | Early to Late (I, IIa, IIb) | High-throughput analysis of thousands of cells; avoids bulk analysis pitfalls [69] | 2.5×10⁵ – 2×10⁶ cells/mL [69] |
| ApoqPCR [70] | Absolute quantitation of apoptotic DNA via ligation-mediated qPCR | Late (IIa, IIb) | Extreme sensitivity; requires sample equivalent to ≤100 cells [70] | As low as 100 cells [70] |
| FLICA Assay with Flow Cytometry [69] | Fluorochrome-labeled inhibitors bind active caspases | Early (I) | Identifies early commitment to death before morphological changes [69] | 2.5×10⁵ – 2×10⁶ cells/mL [69] |
| Mitochondrial Potential Assay (TMRM) [69] | Fluorescent probe accumulation proportional to Δψm | Early (I) | Sensitive marker of early apoptotic events [69] | 2.5×10⁵ – 2×10⁶ cells/mL [69] |
| Western Blot (Antibody Cocktails) [57] | Detects cleavage of specific proteins (e.g., caspases, PARP) | Middle to Late (IIa, IIb) | Multiplexing with antibody cocktails maximizes information from minimal sample [57] | Varies with protein abundance |
Detailed Experimental Protocols
Highly Sensitive DNA Fragmentation Analysis by ApoqPCR
ApoqPCR represents a significant advancement for quantifying apoptosis in minute samples or archived materials, as it provides an absolute measurement of apoptotic DNA with a 1000-fold linear dynamic range and sensitivity for samples equivalent to 100 cells or less [70].
Protocol Steps:
- gDNA Purification: Isolate genomic DNA using mini-columns designed to purify nucleic acid fragments from <200 bp to >50 kbp. Elute in a low-EDTA buffer (e.g., 10 mM Tris–HCl pH 8.5, 0.5 mM EDTA) [70].
- Oligonucleotide Annealing/Ligation:
- Prepare an annealing/ligation reaction containing your test sample gDNA (up to 200 ng), oligonucleotides DHApo1 (24-mer) and DHApo2 (12-mer), and 5x ligation buffer with polyethylene glycol [70].
- Anneal oligonucleotides by stepwise cooling from 55°C to 15°C to form blunt-ended, partially double-stranded linkers.
- Pause at 10°C, add T4 DNA ligase, and continue incubation at 16°C for 16 hours to ligate linkers to apoptotic DNA fragments [70].
- qPCR Amplification and Quantification:
- Use a portion of the ligated reaction in a triplicate qPCR assay.
- Quantify the amount of apoptotic DNA by comparing results to a standard curve generated from serial dilutions of "completely apoptotic" DNA, the concentration of which is determined spectrophotometrically [70].
Multiparameter Flow Cytometry for Early Apoptosis Detection
Flow cytometry is a powerful platform for this application, enabling multiparameter measurements and single-cell analysis, which avoids the sensitivity problems of traditional bulk techniques [69]. The following workflow diagram illustrates a protocol for staining and analysis.
Diagram Title: Flow Cytometry Workflow for Caspase & Membrane Integrity
Protocol Steps (FLICA with Propidium Iodide):
- Cell Preparation: Collect cell suspension (2.5×10⁵ – 2×10⁶ cells/mL) in a FACS tube, centrifuge, and wash the pellet with 1x PBS [69].
- FLICA Staining: Resuspend the cell pellet in PBS and add the FLICA working solution. Incubate for 60 minutes at +37°C, protected from light, gently agitating every 20 minutes [69].
- Washing: Add 2 mL of PBS, centrifuge, and discard the supernatant to remove unbound FLICA reagent. Repeat this wash step [69].
- Viability Staining: Resuspend the cell pellet in a propidium iodide (PI) staining mix. Incubate for 3-5 minutes, then add 500 µL of PBS and keep samples on ice [69].
- Analysis: Analyze immediately on a flow cytometer using 488 nm excitation. FLICA fluorescence (caspase activity) and PI fluorescence (membrane integrity) are collected in appropriate channels, allowing for the discrimination of viable (FLICA-/PI-), early apoptotic (FLICA+/PI-), and late apoptotic/necrotic (FLICA+/PI+) cells [69].
Western Blotting with Antibody Cocktails for Multiplexed Analysis
Using pre-mixed apoptosis antibody cocktails can streamline the detection of multiple apoptotic markers from a single, limited sample, saving time and resources while improving accuracy [57].
Protocol Steps:
- Sample Lysis and Quantification: Prepare cell lysates using appropriate lysis buffers containing protease inhibitors. Perform precise protein quantification to ensure equal loading across all samples [57].
- Electrophoresis and Transfer: Separate proteins by SDS-PAGE and transfer to a membrane [57].
- Multiplexed Immunoblotting:
- Block the membrane to prevent non-specific antibody binding.
- Incubate with a primary apoptosis antibody cocktail (e.g., targeting pro/p17-caspase-3, cleaved PARP1, and a loading control like muscle actin)[ccitation:5].
- After washing, incubate with appropriate secondary antibodies conjugated to detection enzymes or fluorophores [57].
- Visualization and Analysis: Detect signals using chemiluminescence or fluorescence. Use densitometry software (e.g., ImageJ) to quantify band intensities. Critical analysis includes calculating the ratio of cleaved to total protein (e.g., cleaved caspase-3 to total caspase-3) and normalizing to a housekeeping protein (e.g., β-actin) to account for loading variations [57].
The Scientist's Toolkit: Key Research Reagents
Selecting the right reagents is fundamental to the success of these sensitive assays. The following table details essential materials and their functions.
Table 2: Essential Reagents for Apoptosis Detection in Challenging Samples
| Reagent / Assay | Specific Example | Function / Target | Key Consideration for Small Populations |
|---|---|---|---|
| FLICA Reagents [69] | FAM-VAD-FMK (Poly-caspase) | Irreversibly binds to active caspases within live cells. | Allows detection of early apoptotic commitment before plasma membrane permeability, conserving rare cell samples. |
| Mitopotential Dyes [69] | TMRM (Tetramethylrhodamine methyl ester) | Fluorescent cationic probe that accumulates in energized mitochondria; loss of signal indicates Δψm dissipation. | A sensitive marker of very early apoptotic events. The pH of the dye solution must be consistent to avoid artifacts [14]. |
| Annexin V Conjugates [69] | Annexin V-FITC / Annexin V-APC | Binds to phosphatidylserine (PS) exposed on the outer leaflet of the plasma membrane. | Must be performed in the presence of calcium and is typically used with PI to distinguish early (Annexin V+/PI-) from late (Annexin V+/PI+) stages [69]. |
| Apoptosis Antibody Cocktails [57] | ab136812 (Caspase-3, Cleaved PARP, Actin) | Pre-mixed antibodies for simultaneous detection of multiple apoptotic markers in a single western blot. | Maximizes information and conserves precious sample by detecting several key proteins from one gel. |
| Cell Permeability Dyes [69] | Propidium Iodide (PI) | DNA-binding dye that is excluded by intact plasma membranes; stains cells with compromised membranes. | Critical for differentiating apoptotic from necrotic cells in flow cytometry. Handle with care as it is a potential mutagen [69]. |
| qPCR Reagents for ApoqPCR [70] | DHApo1 & DHApo2 Oligonucleotides, T4 DNA Ligase | Enzymes and primers for the specific ligation and amplification of apoptotic DNA fragments. | Enables absolute quantitation of apoptosis from minimal cell inputs, down to hundreds of cells. |
Pathway Context: Connecting Detection to Biology
Understanding the molecular pathways of apoptosis provides context for interpreting results from the aforementioned techniques. A key pathway is the intrinsic (mitochondrial) pathway, which is regulated by BCL-2 family proteins. In this pathway, activation of pro-apoptotic proteins like BAK and BAX leads to mitochondrial outer membrane permeabilization (MOMP), releasing cytochrome c and other factors that activate caspases, the key executioners of apoptosis [71] [72]. The following diagram illustrates this pathway and the points where different detection methods act.
Diagram Title: Apoptosis Detection Methods Mapped to Intrinsic Pathway
Accurately detecting apoptosis in small or rapidly cleared cell populations demands a strategic approach that prioritizes sensitivity, specificity, and the efficient use of sample material. No single method is universally superior; the choice depends on the biological question, the apoptotic phase of interest, and practical laboratory constraints. By leveraging quantitative PCR-based DNA fragmentation analysis, multiparameter flow cytometry for early event detection, and multiplexed immunoblotting, researchers can effectively overcome the challenges posed by limited sample availability and the rapid, efficient nature of apoptotic cell clearance.
Optimizing Sample Preparation and Staining Protocols for Clear Morphology
The detection of apoptosis is fundamentally linked to the observation of distinct morphological features, which represent a cornerstone for understanding tissue differentiation, organ development, aging, and the elimination of mutant cells [14]. These morphological changes occur in a phased manner, and their clear visualization is essential for accurate interpretation of experimental results in fields ranging from basic cell biology to preclinical drug development [14] [18]. For researchers and drug development professionals, selecting and optimizing the correct preparation and staining protocol is not merely a technical step but a strategic decision that directly impacts data quality and biological insight. This guide provides a detailed framework for optimizing these protocols, specifically framed within the context of researching the morphological features of apoptosis Phases I, IIa, and IIb. A purpose-dependent approach is crucial, as the choice of method must align with the specific apoptotic phase and research question being investigated [14].
Morphological Hallmarks of Apoptosis Phases I, IIa, and IIb
Apoptosis progresses through a series of morphologically distinct phases, each characterized by specific cellular and nuclear changes. Understanding these hallmarks is a prerequisite for selecting the appropriate detection method.
Phase I is marked by the contraction of the cell. The apoptotic cell shrinks and acquires a dense cytoplasm, a consequence of decreased water content and increased eosinophilia. Microvilli on the cell surface disappear, and critically, the cell separates from the surrounding normal cell population [14]. At the ultrastructural level, transmission electron microscopy reveals the appearance of many vacuoles, a process known as cavitation [14].
Phase IIa involves dramatic nuclear alterations. Chromatin undergoes condensation, becoming dense masses (pyknosis) or assembling on the inner nuclear membrane (chromatin margination). Subsequently, the nuclei are broken into fragments (fragmentation) [14]. Electron microscopy is particularly effective for observing these changes.
Phase IIb represents the final morphological stage. The cytoskeleton degrades, causing invaginations in the cell membrane, or sprouting and displacement. This leads to the formation of membrane-coated vesicles containing cytoplasmic membrane, nuclear debris, and organelle components. These vesicles are transformed into small bodies known as apoptotic bodies, which are arguably the most important morphological markers of apoptosis [14]. The integrity of the plasma membrane is generally maintained, preventing the contents from being released and triggering a peripheral inflammatory response.
Table 1: Morphological Characteristics of Apoptosis Phases I, IIa, and IIb
| Apoptosis Phase | Key Cellular Morphology | Key Nuclear Morphology | Primary Observation Method |
|---|---|---|---|
| Phase I | Cell shrinkage, dense cytoplasm, increased eosinophilia, loss of surface microvilli, cavitation [14]. | --- | Electron Microscopy [14] |
| Phase IIa | --- | Chromatin condensation (pyknosis), margination to nuclear membrane, nuclear fragmentation [14]. | Electron Microscopy, Fluorescence/Confocal Microscopy [14] |
| Phase IIb | Membrane blebbing, cytoskeleton disintegration, formation of apoptotic bodies [14]. | Nuclear debris packaged into apoptotic bodies [14]. | Light Microscopy, Electron Microscopy, Fluorescence/Confocal Microscopy [14] |
Detection Methodologies for Morphological Analysis
A variety of techniques are available for visualizing the morphological features of apoptosis, each with distinct advantages, disadvantages, and applicability to the different phases.
Light Microscopy
After staining with common dyes like hematoxylin and eosin (HE), Giemsa, or Wright's stain, light microscopy can reveal cell shrinking, rounding, and shedding of nuclei, as well as the presence of apoptotic bodies [14]. It is mainly suitable for observing Phase IIb apoptosis, where apoptotic bodies are formed [14]. A significant limitation is that phagocytosis of apoptotic cells is very effective and rapid, meaning apoptosis in a small area is not easily recognized via light microscopy [14].
Electron Microscopy
Considered the gold standard for detailed morphological assessment, electron microscopy after uranyl acetate-lead citrate staining can reveal the ultra-morphological changes across all phases of apoptosis (I, IIa, and IIb) [14]. It provides the finest detail, including vacuolation in Phase I, highly condensed and marginalized chromatin in Phase IIa, and nuclear fragmentation in Phase IIb [14]. However, not all cells exhibit typical morphological features during apoptosis, and its inability to process large sample volumes can be a constraint [14].
Fluorescence/Confocal Microscopy
This technique uses DNA-binding fluorescent dyes like Hoechst 33,342, acridine orange (AO), or DAPI to indirectly reveal the condition of the nucleus and chromatin [14]. The intensity and distribution of the fluorescence signals determine the occurrence of apoptosis. For instance, condensed or fragmented chromatin will appear as brighter, punctate spots. It is mainly suitable for observing Phase IIb apoptosis and, like light microscopy, may miss apoptosis in small areas [14].
Table 2: Comparison of Morphological Detection Methods for Apoptosis
| Method | Key Advantages | Key Disadvantages | Optimal Apoptosis Phase |
|---|---|---|---|
| Light Microscopy | Simple, convenient, intuitive, storable specimens [14]. | Misses small areas of apoptosis; mainly for late stages [14]. | Phase IIb [14] |
| Electron Microscopy | Reveals typical ultra-morphology and structure; suitable for all phases [14]. | Cannot rule out apoptosis without typical features; not high-throughput [14]. | Phases I, IIa, IIb [14] |
| Fluorescence/Confocal Microscopy | Directly reveals nuclear and chromatin changes; high-resolution 3D imaging [14]. | Misses small areas of apoptosis; mainly for late stages [14]. | Phase IIb [14] |
Optimized Staining Protocols for Clear Morphology
Annexin V/Propidium Iodide (PI) Staining for Flow Cytometry
The Annexin V/PI assay is a powerful biochemical tool that complements pure morphology by identifying early membrane changes and distinguishing them from late-stage membrane rupture. The protocol below ensures accurate quantification of cell populations.
Materials Needed:
- Cells: Cultured cells or cell suspension (e.g., MDA-MB-231 breast cancer cell line) [73].
- Fluorochrome-conjugated Annexin V (e.g., FITC, APC) [73] [74].
- Propidium Iodide (PI) solution (1 μg/mL) [73].
- 1X Binding Buffer (PBS with 25 mM CaCl₂; critical: avoid EDTA) [73] [74].
- Phosphate Buffered Saline (PBS), without calcium and magnesium [73].
- Flow cytometer equipped with 488 nm blue and 633 nm red lasers [73].
Step-by-Step Protocol:
- Induce Apoptosis and Harvest Cells: Treat cells with an apoptosis-inducing agent (e.g., 1 μM doxorubicin for 48 hours) [73]. For adherent cells, harvest by gentle trypsinization and combine with the supernatant to collect any detached (dead) cells [73] [75]. For suspension cells, collect directly.
- Wash and Resuspend: Wash cells once with PBS and once with 1X Binding Buffer by centrifugation at 300×g for 5 minutes [73] [74]. Resuspend the cell pellet in 1X Binding Buffer at a concentration of 1 x 10⁶ cells/mL [74].
- Stain Cells: Aliquot 100 μL of cell suspension (1 x 10⁵ cells) into a flow tube. Add 5 μL of Annexin V conjugate and 5 μL of PI solution [74]. Gently mix the tubes.
- Incubate: Incubate at room temperature for 15 minutes in the dark [73] [74].
- Analyze: Add 400 μL of 1X Binding Buffer to each tube and analyze promptly by flow cytometry (within 1 hour) [75] [74]. Keep samples on ice if analysis is delayed.
Interpretation of Results:
- Viable Cells: Annexin V-FITC negative, PI negative.
- Early Apoptotic Cells: Annexin V-FITC positive, PI negative (PS externalized, membrane intact).
- Late Apoptotic Cells: Annexin V-FITC positive, PI positive (PS externalized, membrane compromised).
- Necrotic Cells: Annexin V-FITC negative, PI positive [73] [76].
Fluorescence Staining for Nuclear Morphology (Hoechst/DAPI)
This protocol is ideal for visualizing the nuclear changes characteristic of Phases IIa and IIb, such as chromatin condensation and fragmentation.
Materials Needed:
- Cells grown on glass coverslips or in suspension.
- Fixative (e.g., 4% Paraformaldehyde in PBS) [75].
- Permeabilization buffer (e.g., 0.1% Triton X-100 in 0.1% sodium citrate) [75].
- Fluorescent DNA-binding dye (e.g., Hoechst 33342 or DAPI) [14] [77].
- Mounting medium.
- Fluorescence or confocal microscope.
Step-by-Step Protocol:
- Fix Cells: Wash cells with PBS and fix with 4% paraformaldehyde for 30 minutes at room temperature [75].
- Permeabilize Cells: Wash fixed cells with PBS and incubate with permeabilization buffer for 2 minutes on ice [75].
- Stain Nuclei: Wash cells and incubate with an appropriate concentration of Hoechst 33342 or DAPI for 10-30 minutes at room temperature in the dark [14] [77].
- Mount and Visualize: Wash off excess stain, mount the coverslip onto a glass slide, and visualize under a fluorescence microscope. Apoptotic cells will display brightly stained, condensed, and often fragmented nuclei compared to the diffuse staining of viable cells [14].
Integrating Morphology with Biochemical and Immunological Techniques
A comprehensive apoptosis analysis often requires correlating morphology with biochemical events. Key techniques include:
- DNA Gel Electrophoresis: Detects the classic "ladder" pattern of DNA fragmentation (180-200 bp and multiples) caused by endonuclease activation. It is simple and accurate but lacks cellular localization and is only suitable for middle and late stages of apoptosis with large-scale cell death [14].
- TUNEL Assay (Terminal deoxynucleotidyl transferase dUTP Nick End Labeling): This method uses terminal transferase (TdT) to label the 3'-OH ends of DNA fragments, which are abundant in apoptotic cells. It is relatively sensitive and specific for counting and quantifying apoptotic cells in situ, making it suitable for late-stage apoptosis, though false positives can occur [14] [75]. Modern kits, such as the Click-iT TUNEL assays, offer improved sensitivity and flexibility for multiplexing with other biomarkers [77].
- Caspase Activity Detection: Apoptotic proteases (caspases) are key executors of apoptosis. Advanced real-time approaches use Fluorescence Resonance Energy Transfer (FRET)-based genetically encoded caspase sensors. Cells stably expressing such a probe (e.g., CFP and YFP linked by a DEVD caspase cleavage site) will exhibit a loss of FRET (change in emission ratio) upon caspase activation, providing a dynamic, single-cell readout of apoptosis that can be combined with morphological assessment [18].
Signaling Pathways in Apoptosis
Understanding the signaling cascades that drive the morphological changes provides a deeper context for research. The mechanism of apoptosis is primarily divided into exogenous (death receptor) and endogenous (mitochondrial and endoplasmic reticulum) pathways [14]. The mitochondrial pathway is a key endogenous route.
This diagram illustrates the core mitochondrial pathway. An apoptotic stimulus leads to the inhibition of anti-apoptotic proteins like Bcl-2 [14]. This triggers Mitochondrial Outer Membrane Permeabilization (MOMP), resulting in the release of cytochrome C into the cytosol [14]. Cytochrome C then initiates the assembly of the apoptosome and activation of the caspase cascade [14]. The downstream activation of executioner caspases directly cleaves cellular targets, leading to the sequential morphological changes that define Phases I, IIa, and IIb [14].
The Scientist's Toolkit: Essential Reagents for Apoptosis Morphology Research
Table 3: Key Research Reagent Solutions for Apoptosis Morphology Studies
| Reagent / Kit | Primary Function | Key Application in Apoptosis Research |
|---|---|---|
| Annexin V Conjugates (e.g., FITC, PE, APC) [73] [74] | Binds to externalized phosphatidylserine (PS) in a calcium-dependent manner. | Detection of early apoptotic cells (Phase I/IIa) by flow cytometry or microscopy [78] [76]. |
| Viability Dyes (Propidium Iodide, 7-AAD) [73] [74] [77] | DNA intercalating dyes excluded by intact membranes. | Discrimination of late apoptotic/necrotic cells (membrane compromised) from early apoptotic cells [73] [76]. |
| Nuclear Stains (Hoechst 33342, DAPI) [14] [77] | Bind DNA and label the nucleus. | Visualization of nuclear morphology changes like chromatin condensation and fragmentation (Phase IIa/IIb) [14] [77]. |
| TUNEL Assay Kits [75] [77] | Enzymatically labels 3'-OH ends of fragmented DNA. | In situ detection of late-stage apoptotic cells with DNA fragmentation [14] [77]. |
| Caspase Detection Kits (FRET-based, fluorogenic substrates) [18] | Detects activation of caspase enzymes. | Confirmation of apoptosis via a key biochemical event; allows real-time analysis and distinction from necrosis [18]. |
| Antibodies to Apoptosis Markers (e.g., cleaved PARP, cleaved Caspase-3) [75] [77] | Immunofluorescence detection of cleaved/activated proteins. | Correlates biochemical events (e.g., caspase activation) with morphological changes in fixed cells [77]. |
Optimizing sample preparation and staining protocols is paramount for achieving clear and interpretable morphology in apoptosis research. The choice of method—whether light, electron, or fluorescence microscopy, and whether to integrate it with biochemical assays like Annexin V/PI or TUNEL—must be guided by the specific apoptotic phase of interest and the research objectives. A multifaceted approach that correlates the classic morphological hallmarks of Phases I, IIa, and IIb with underlying biochemical events provides the most robust and insightful data. This structured, purpose-dependent framework empowers researchers and drug developers to generate high-quality, reproducible results, ultimately advancing our understanding of cell death in health and disease.
The Importance of Combining Morphological with Biochemical Methods for Confirmation
Abstract Within the context of a broader thesis on the morphological features of apoptosis phases I, IIa, and IIb, this technical guide underscores the critical necessity of integrating morphological and biochemical analytical techniques. Apoptosis, a programmed and energy-dependent cell death process, is characterized by a cascade of molecular events and distinct morphological stages. Relying on a single detection method risks misclassification, especially given the expanding spectrum of programmed cell death (PCD) pathways with overlapping yet distinct features. This whitepaper provides researchers, scientists, and drug development professionals with a detailed framework for the concurrent application of morphological and biochemical assays, complete with structured data, experimental protocols, and visualization tools to ensure accurate confirmation of apoptotic cell death.
1. Introduction The precise identification of apoptotic cells is paramount in diverse fields, from basic biological research to the development of novel anti-cancer therapeutics. Apoptosis proceeds through tightly regulated phases—conventionally termed Phase I (cell shrinkage), Phase IIa (nuclear condensation), and Phase IIb (formation of apoptotic bodies)—each defined by specific morphological and biochemical hallmarks [14]. However, the discovery of multiple caspase-independent regulated necrosis pathways, such as necroptosis, pyroptosis, and ferroptosis, has complicated the cell death landscape, as these pathways can exhibit morphological features reminiscent of both apoptosis and accidental necrosis [17] [79]. For instance, necroptosis is a regulated form of death that shares the cell swelling and membrane rupture of necrosis but is genetically controlled [17]. This convergence and divergence of phenotypes make it challenging to rely on a single analytical method. Therefore, a multi-parametric approach that combines direct visualization of morphological changes with the detection of specific biochemical signatures is essential for unambiguous confirmation of apoptosis and for distinguishing it from other PCD modalities.
2. Morphological Hallmarks of Apoptosis Phases I, IIa, and IIb The morphological classification of apoptosis provides the initial, visible evidence of cell death execution. These changes are best observed using microscopy techniques, each with its own advantages and limitations for identifying specific phases.
Table 1: Morphological Characteristics and Detection Methods Across Apoptosis Phases
| Apoptosis Phase | Key Morphological Features | Recommended Detection Methods | Technical Considerations |
|---|---|---|---|
| Phase I | Cell shrinkage and rounding; increased cytoplasmic density; disappearance of cell-surface specializations (e.g., microvilli) [14]. | Transmission Electron Microscopy (TEM); Full-field optical coherence tomography (FF-OCT) [14] [20]. | TEM provides ultra-high resolution but requires fixed samples; FF-OCT is label-free and allows for live-cell, 3D monitoring [20]. |
| Phase IIa | Chromatin condensation (pyknosis); margination of condensed chromatin against the nuclear envelope [14]. | Fluorescence microscopy (Hoechst 33342, DAPI); TEM [14] [80]. | Fluorescent DNA dyes like Hoechst 33342 show increased fluorescence intensity and nuclear condensation in apoptotic cells, allowing distinction from healthy and necrotic cells [80]. |
| Phase IIb | Nuclear fragmentation (karyorrhexis); membrane blebbing; formation of apoptotic bodies containing nuclear debris and organelles [14]. | Light microscopy (HE, Giemsa staining); Fluorescence/Confocal microscopy; FF-OCT [14] [20]. | Light microscopy is simple but may miss early stages; FF-OCT can visualize dynamic membrane blebbing and filopodia reorganization in real-time without labels [14] [20]. |
3. Biochemical Hallmarks and Corresponding Detection Assays Biochemical events run in parallel to morphological changes, providing specific, often quantifiable, targets for apoptosis confirmation. Key biomarkers include phosphatidylserine externalization, caspase activation, and DNA fragmentation.
Table 2: Key Biochemical Biomarkers of Apoptosis and Their Detection
| Biomarker | Biochemical Event | Detection Assays | Stage Detected |
|---|---|---|---|
| Phosphatidylserine (PS) Externalization | Translocation of PS from the inner to the outer leaflet of the plasma membrane, an "eat-me" signal [81]. | Annexin V staining (often combined with a viability dye like PI) measured by flow cytometry or fluorescence microscopy [12] [81]. | Early Apoptosis |
| Caspase Activation | Proteolytic cleavage and activation of executioner caspases (e.g., Caspase-3, -7) [17] [82]. | Fluorometric/Colorimetric caspase activity assays; Western blot for cleaved caspases [82] [81]. | Early-to-Mid Apoptosis |
| DNA Fragmentation | Cleavage of DNA into oligonucleosomal fragments (180-200 bp) by activated endonucleases [14] [82]. | TUNEL assay; DNA gel electrophoresis (DNA laddering) [14] [82]. | Late Apoptosis |
| Cytochrome c Release | Release from mitochondria into the cytosol following mitochondrial outer membrane permeabilization (MOMP) [17] [81]. | ELISA of cytosolic fractions; immunofluorescence [12] [81]. | Mid Apoptosis |
| Loss of Mitochondrial Membrane Potential (ΔΨm) | Disruption of the electrochemical gradient across the mitochondrial inner membrane [14] [81]. | Fluorescent dyes (e.g., JC-1, TMRM) measured by flow cytometry or fluorescence microscopy [14] [81]. | Early Apoptosis |
4. Experimental Protocols for Combined Analysis To ensure robust confirmation, the following protocols outline methods that can be performed sequentially or in parallel on the same cell population.
4.1. Protocol: Combined Annexin V/Propidium Iodide (PI) Staining and Nuclear Morphology Analysis This protocol allows for the simultaneous assessment of an early biochemical marker (PS externalization) and late-stage morphological nuclear changes.
- Cell Staining: Harvest cells (adherent cells should be gently trypsinized). Resuspend cell pellet (~1x10⁶ cells) in 100 µL of binding buffer.
- Add Fluorochromes: Add Annexin V conjugated to a fluorophore (e.g., FITC or Cy3) and a membrane-impermeant DNA dye like Propidium Iodide (PI) [81]. Incubate for 15 minutes in the dark at room temperature.
- Flow Cytometry Analysis: Add additional binding buffer and analyze by flow cytometry. Quadrant analysis distinguishes:
- Annexin V-/PI-: Viable, non-apoptotic cells.
- Annexin V+/PI-: Early apoptotic cells (PS externalized, membrane intact).
- Annexin V+/PI+: Late apoptotic or necrotic cells (membrane integrity lost).
- Fluorescence Microscopy: After staining, pellet cells and resuspend in a small volume. Place on a microscope slide and add a DNA-staining dye like Hoechst 33342 (2 µg/mL) for 10 minutes [80].
- Image Acquisition and Analysis: Visualize under a fluorescence microscope. Correlate Annexin V positivity with nuclear morphology: viable cells (Annexin V-, Hoechst-diffuse), early apoptotic (Annexin V+, Hoechst-condensed), and late apoptotic/necrotic (Annexin V+/PI+, Hoechst-condensed/fragmented).
4.2. Protocol: Caspase Activity Assay Coupled with FF-OCT Morphological Imaging This combination validates the activation of a central biochemical effector with high-resolution, label-free 3D morphology.
- Induction and Sampling: Induce apoptosis in cultured cells (e.g., HeLa cells with 5 µM doxorubicin) [20]. At various time points, harvest a subset of cells for biochemical assay.
- Caspase-3/7 Activity Assay: Lyse the harvested cells. Incubate cell lysates with a caspase-specific substrate (e.g., DEVD-peptide conjugated to a fluorophore or chromophore). Measure the cleavage product using a fluorometer or spectrophotometer to quantify caspase activity [81].
- FF-OCT Live-Cell Imaging: For the remaining cells, perform real-time imaging using a custom-built FF-OCT system [20]. This technique provides en face (x-y) cross-sectional and 3D surface topography maps without labels.
- Correlative Data Interpretation: Correlate the kinetic data from the caspase activity assay with the morphological changes observed via FF-OCT, such as the emergence of echinoid spines, membrane blebbing, and cell contraction, which are characteristic of apoptosis [20].
5. The Scientist's Toolkit: Essential Reagent Solutions A selection of key reagents and kits for apoptosis detection is summarized below.
Table 3: Research Reagent Solutions for Apoptosis Detection
| Reagent / Kit | Function / Target | Application Notes |
|---|---|---|
| Annexin V Conjugates | Binds to externalized Phosphatidylserine (PS) [81]. | Critical for flow cytometry and microscopy to detect early apoptosis. Must be used with a viability dye (e.g., PI) to assess membrane integrity. |
| Caspase Activity Assay Kits | Measure the proteolytic activity of specific caspases (e.g., Caspase-3) using colorimetric or fluorometric substrates [81]. | Provides quantitative data on a central apoptotic event. Distinguishes between early (initiator caspases) and late (executioner caspases) stages. |
| Hoechst 33342 / DAPI | Cell-permeable DNA dyes that stain the nucleus. Fluorescence intensifies upon chromatin condensation [14] [80]. | A simple and direct method for visualizing nuclear morphology changes characteristic of Phase IIa and IIb apoptosis by fluorescence microscopy. |
| Mitochondrial Membrane Potential Dyes (e.g., JC-1) | Accumulate in mitochondria in a potential-dependent manner, emitting different fluorescence colors based on ΔΨm status [14] [81]. | Useful for detecting early intrinsic apoptosis. A shift from red (aggregate) to green (monomer) fluorescence indicates loss of ΔΨm. |
| TUNEL Assay Kits | Labels the 3'-OH ends of fragmented DNA in situ [14] [82]. | Highly specific for detecting late-stage apoptosis (DNA fragmentation). Can be used for histology sections, fixed cells, and flow cytometry. |
| Cytochrome c ELISA Kits | Quantifies cytochrome c release from mitochondria into the cytosol [12] [81]. | Requires subcellular fractionation to separate mitochondrial and cytosolic components. Confirms activation of the intrinsic apoptotic pathway. |
6. Conclusion In the rigorous landscape of cell death research, particularly for a thesis focused on the nuanced stages of apoptosis, reliance on a single analytical method is insufficient. The morphological features of Phases I, IIa, and IIb provide a visible narrative of cellular demise, while biochemical assays offer precise, mechanistic validation of the underlying molecular machinery. Their combined use is not merely recommended but is essential for definitive confirmation, enabling researchers to distinguish apoptosis from other programmed necrosis pathways with high confidence. This integrated strategy is fundamental for advancing our understanding of cell death in development, homeostasis, and disease, and is critical for the accurate evaluation of novel therapeutic agents in drug development pipelines.
Integrating Morphology with Molecular Biology for a Complete Apoptosis Profile
Apoptosis, or programmed cell death, is a fundamental physiological process crucial for maintaining tissue homeostasis, embryonic development, and eliminating damaged or potentially harmful cells [83] [57]. It is a tightly regulated mechanism characterized by distinct morphological changes and biochemical events. In pathological contexts, particularly in cancer research, defects in apoptotic pathways allow damaged cells to survive and proliferate, making the modulation of these pathways a primary therapeutic goal [83] [84] [57]. Accurate detection and validation of apoptosis are therefore paramount in molecular biology, disease modeling, and drug screening.
This technical guide focuses on the critical cross-validation of classical morphological phases of apoptosis with two established biochemical assays: the DNA laddering assay and the Terminal deoxynucleotidyl transferase dUTP Nick-End Labeling (TUNEL) assay. The core thesis is that a robust understanding of apoptosis requires correlating the visible, phase-based morphological progression of a dying cell with the hallmark biochemical signature of internucleosomal DNA fragmentation. This multi-parameter approach is essential for researchers and drug development professionals to confidently interpret cell death mechanisms in response to various stimuli.
The Morphological Phases of Apoptosis
Apoptosis unfolds through a sequence of three primary morphological phases, each defined by specific cellular alterations. These phases provide a visual framework for identifying dying cells and contextualizing biochemical data.
- Phase I (Early Phase): The cell initiates a self-destruction program. The earliest visible signs include cell shrinkage and a reduction in water content, leading to a denser cytoplasm. The cell begins to detach from its extracellular matrix and neighboring cells, and surface structures like microvilli are lost [57].
- Phase IIa (Middle Phase): This phase is dominated by profound nuclear changes. The chromatin within the nucleus undergoes condensation, forming dense, marginalized masses along the inner nuclear membrane. Subsequently, the nucleus itself fragments into discrete bodies [57].
- Phase IIb (Late Phase): The cell enters its final stage of disintegration. The cytoskeleton degrades, and the cell membrane blebs, eventually pinching off to form membrane-bound vesicles known as apoptotic bodies. These bodies contain nuclear debris and cytoplasmic components and are swiftly cleared by phagocytic cells, preventing an inflammatory response [57].
Table 1: Morphological Phases of Apoptosis and Key Characteristics
| Morphological Phase | Key Cellular and Nuclear Characteristics |
|---|---|
| Phase I (Early) | Cell shrinkage, reduced water content, increased cytoplasmic density, detachment from substrate, loss of microvilli [57]. |
| Phase IIa (Middle) | Chromatin condensation, nuclear fragmentation, formation of dense chromatin masses [57]. |
| Phase IIb (Late) | Overall fragmentation into apoptotic bodies, membrane blebbing, degradation of cytoskeletal structures [57]. |
Biochemical Hallmarks and Detection Assays
The morphological changes during apoptosis are driven by a conserved biochemical cascade. A key hallmark is the activation of endogenous endonucleases that cleave nuclear DNA at the linker regions between nucleosomes, generating fragments of ~180-200 base pairs and their multiples [83]. This specific DNA fragmentation is the molecular basis for the DNA laddering and TUNEL assays.
DNA Laddering Assay
The DNA laddering assay is a classic, cost-effective method for detecting the internucleosomal DNA fragmentation characteristic of apoptosis. It involves extracting DNA from a cell population, separating the fragments via agarose gel electrophoresis, and visualizing a distinctive "ladder" pattern, which contrasts with the smeared pattern of necrotic DNA [83].
Improved Experimental Protocol for DNA Ladder Assay [83]:
- Cell Culture and Apoptosis Induction: Culture cells (e.g., NIH-3T3) and induce apoptosis using an appropriate stimulus (e.g., 500 µM H₂O₂ for 48 hours).
- DNA Extraction:
- Critical Step: Collect culture media containing detached (apoptotic) cells and centrifuge (5,000 rpm for 5 minutes). Retain the pellet.
- Add lysis buffer (e.g., 2% buffer containing Tris-HCl, EDTA, NaCl, C-TAB) to the culture vessel to harvest remaining adherent cells. Combine this lysate with the pellet from the previous step.
- Incubate the combined lysate at 65°C for 5 minutes.
- Cool to room temperature, add an equal volume of chloroform-isoamyl alcohol, and centrifuge (12,000 rpm for 5 minutes).
- Transfer the upper aqueous phase to a new tube. Add an equal volume of cold isopropanol to precipitate the DNA. Mix gently by inversion.
- Centrifuge (12,000 rpm for 5 minutes), discard the supernatant, and air-dry the DNA pellet for 30 minutes.
- Resuspend the purified DNA in nuclease-free water and quantify using a spectrophotometer.
- Gel Electrophoresis and Visualization: Load the DNA samples onto a 1.5% agarose gel containing a DNA stain (e.g., SYBR-Safe). Perform electrophoresis and visualize the DNA ladder pattern using an ultraviolet gel documentation system.
TUNEL Assay
The TUNEL (TdT dUTP Nick-End Labeling) assay is a highly sensitive technique that detects DNA strand breaks, a late-stage feature of apoptosis, by enzymatically labeling the 3'-OH ends of fragmented DNA. It allows for the in situ detection of apoptotic cells within a tissue section or cell culture and is amenable to quantification by flow cytometry [85].
Cross-Validating Morphology with Biochemistry
The strength of a multi-faceted approach lies in correlating data from different methodologies. The table below provides a systematic summary of how the morphological phases align with the expected outcomes from DNA laddering and TUNEL assays, as well as other confirmatory techniques.
Table 2: Cross-Validation of Apoptosis: Morphological Phases and Assay Outcomes
| Morphological Phase | DNA Ladder Assay Result | TUNEL Assay Result | Additional Confirmatory Assays |
|---|---|---|---|
| Phase I (Early) | Usually negative | Usually negative | Flow Cytometry (Annexin V/PI): Annexin V-positive, PI-negative (indicating phosphatidylserine exposure) [83]. |
| Phase IIa (Middle) | Intermittent / Weak Ladder: Onset of internucleosomal cleavage may be detectable. | Positive: Chromatin condensation and early fragmentation generate DNA breaks for labeling [85]. | DAPI Staining: Visible chromatin condensation and nuclear fragmentation under fluorescence microscopy [83]. Western Blot: Initial cleavage/activation of caspases and PARP [57]. |
| Phase IIb (Late) | Strong, Distinct Ladder: Widespread DNA fragmentation yields a clear banding pattern [83]. | Strongly Positive: Extensive DNA fragmentation leads to intense labeling [85]. | Flow Cytometry (Annexin V/PI): Annexin V and PI-positive (loss of membrane integrity). Western Blot: Prominent levels of cleaved caspases (e.g., Caspase-3) and cleaved PARP [57]. Quantitative Phase Imaging (QPI): Records dynamic changes like cell shrinkage and membrane blebbing [58]. |
Integrated Signaling Pathways and Experimental Workflow
The morphological and biochemical events of apoptosis are executed via specific signaling pathways. The extrinsic pathway is triggered by external death signals, while the intrinsic pathway is initiated by internal cellular stress. Both converge on the activation of caspases, which mediate the cellular dismantling process [57]. The following diagram and workflow illustrate the connection between these molecular events and the experimental methods used for their detection.
Diagram 1: Apoptosis pathways and detection methods.
The logical workflow for a comprehensive apoptosis study, integrating the principles above, is outlined below.
Diagram 2: Experimental workflow for apoptosis analysis.
The Scientist's Toolkit: Essential Reagents and Materials
Successful execution of apoptosis assays requires specific reagents and materials. The following table details key components for the featured experiments.
Table 3: Research Reagent Solutions for Apoptosis Detection
| Item / Reagent | Function / Application | Specific Examples / Notes |
|---|---|---|
| Cell Lines | Model systems for in vitro apoptosis research. | NIH-3T3 [83], DU145, LNCaP, PNT1A [58]. |
| Apoptosis Inducers | Chemical triggers to initiate programmed cell death. | H₂O₂ (500 µM) [83], Doxorubicin (0.1 µM) [58], Staurosporine (0.5 µM) [58]. |
| DNA Ladder Assay Kits | Provide optimized buffers and reagents for DNA fragmentation analysis. | Components include Lysis Buffer (Tris-HCl, EDTA, NaCl, C-TAB), Chloroform-Isoamyl Alcohol, Cold Isopropanol [83]. |
| TUNEL Assay Kits | Enable fluorescent or colorimetric detection of DNA strand breaks in situ. | Kits include Terminal Deoxynucleotidyl Transferase (TdT) and labeled dUTP for end-labeling [85]. |
| Antibodies for Western Blot | Detect specific protein markers and their cleavage products. | Antibodies against Cleaved Caspase-3, Cleaved PARP, Bax, Bcl-2 [57]. Apoptosis antibody cocktails increase efficiency [57]. |
| Flow Cytometry Reagents | Allow quantification of early/late apoptotic populations. | Annexin V-FITC (binds phosphatidylserine), Propidium Iodide (PI) stains DNA in dead cells [83]. |
| Fluorescent Stains | Visualize nuclear morphology and other cellular changes. | DAPI (stains DNA, shows condensation) [83], Hoechst 33342 [58], CellEvent Caspase-3/7 reagent [58]. |
Advanced Techniques and Future Perspectives
While DNA laddering and TUNEL are foundational, the field is advancing with more dynamic and quantitative technologies. Quantitative Phase Imaging (QPI) is a powerful label-free method that allows time-lapse observation of subtle changes in cell mass distribution, density, and morphology, enabling the distinction between apoptosis and lytic cell death based on dynamical features [58]. Furthermore, FRET-based genetically encoded sensors permit real-time, single-cell analysis of caspase activation, providing unparalleled resolution for discriminating apoptosis from necrosis in live cells [84]. The integration of these advanced techniques with classical assays will continue to enhance the precision and depth of apoptosis research in drug discovery and mechanistic studies.
Comparing Morphological Features Across Different Cell Types and Tissues
Apoptosis, or programmed cell death, is a fundamental biological process crucial for maintaining tissue homeostasis, eliminating damaged, unnecessary, or potentially harmful cells in a controlled and organized manner [57]. Unlike necrosis, which is an uncontrolled, inflammatory form of cell death, apoptosis is characterized by a clean and non-inflammatory demise where the dying cell is packaged into small, membrane-bound fragments called apoptotic bodies for removal by immune cells [57]. The morphological progression of apoptosis is classically divided into three distinct phases: Phase I (early), Phase IIa (middle), and Phase IIb (late) [14] [57]. Each phase exhibits specific, recognizable morphological features that can be identified using various detection methodologies. Understanding these morphological changes across different cell types and tissues is paramount for researchers and drug development professionals, as dysregulation of apoptosis is implicated in a wide range of diseases, including cancer and neurodegenerative disorders [57]. This guide provides an in-depth technical comparison of these morphological features and details the experimental protocols for their detection.
Morphological Characteristics of Apoptosis Phases
The following table summarizes the key morphological characteristics associated with each phase of apoptosis, providing a structured comparison for identification purposes.
Table 1: Morphological Characteristics of Apoptosis Phases I, IIa, and IIb
| Apoptosis Phase | Cellular and Nuclear Morphology | Key Observable Features | Recommended Detection Methods |
|---|---|---|---|
| Phase I (Early) | Cell shrinkage; acquisition of a dense cytoplasm; decreased water content; increased eosinophilia; disappearance of cell surface microvilli; separation from surrounding normal cells [14] [57]. | Cell contraction and increased cytoplasmic density. | Electron microscopy (for ultrastructural changes, including vacuole/cavitation formation) [14]. Analysis of mitochondrial membrane potential using fluorescent dyes [14]. |
| Phase IIa (Middle) | Chromatin condensation (pyknosis) forming dense masses, or assembly on the inner nuclear membrane (chromatin margination); initial nuclear fragmentation [14] [57]. | Highly condensed and marginalized chromatin; nuclear breakup begins. | Fluorescence/confocal microscopy with DNA stains (Hoechst, DAPI, AO) [14]. Electron microscopy [14]. |
| Phase IIb (Late) | Degradation of the cytoskeleton; invaginations in the cell membrane, sprouting, and displacement; formation of membrane-coated vesicles (apoptotic bodies) containing nuclear debris, cytoplasmic membrane, and organelle components [14] [57]. | Formation of apoptotic bodies; membrane blebbing. | Light microscopy (HE, Giemsa, Wright's staining) [14]. Fluorescence/confocal microscopy [14]. DNA gel electrophoresis (for DNA laddering) [14]. TUNEL assay [14]. |
Methodologies for Detecting Morphological Changes
A combination of techniques is employed to visualize the structural alterations during apoptosis, each with unique advantages and applications.
Conventional Microscopy Techniques
Light Microscopy is suitable for observing Phase IIb apoptosis, where features like cell rounding, nuclear shedding, and apoptotic bodies are visible after staining with hematoxylin and eosin (HE), Giemsa, or Wright's stains [14]. Its advantages include simplicity and the ability to create storable specimens. However, apoptosis in small areas is easily missed as apoptotic cells are rapidly phagocytosed [14].
Electron Microscopy, particularly transmission electron microscopy (TEM) after uranyl acetate-lead citrate staining, reveals the ultra-morphological changes across all three apoptotic phases [14]. It can identify cell shrinkage, concentrated cytoplasm, surface protrusions in Phase I, highly condensed and marginalized chromatin in Phase IIa, and nuclear fragmentation into apoptotic bodies in Phase IIb [14]. While it provides definitive typical apoptotic morphology, the absence of these features does not rule out apoptosis, necessitating complementary detection methods [14].
Fluorescence or Confocal Microscopy directly reveals nuclear changes when combined with DNA-binding fluorescent dyes like Hoechst 33342, Acridine Orange (AO), or DAPI [14]. The intensity and distribution of fluorescence signals indicate nuclear and chromatin conditions, making it particularly suitable for observing the nuclear fragmentation of Phase IIb [14]. However, similar to light microscopy, it is not ideal for detecting small areas of apoptosis [14].
Advanced Label-Free Imaging
Full-Field Optical Coherence Tomography (FF-OCT) is an emerging, high-resolution, interferometric imaging technique that enables label-free, non-invasive, and real-time visualization of cellular structural changes, overcoming limitations of staining-based methods [20]. A recent study used a custom-built time-domain FF-OCT system to monitor doxorubicin-induced apoptosis in HeLa cells, successfully capturing characteristic features such as:
- Echinoid spine formation
- Cell contraction
- Membrane blebbing
- Filopodia reorganization [20]
FF-OCT's use of a broadband light source with high-magnification objectives allows for subcellular 3D imaging, enabling the reconstruction and quantitative analysis of cell surface topography and internal organelle distributions throughout the apoptotic process [20]. This makes it a powerful platform for distinguishing cell death pathways and assessing dynamic cellular states without the need for labels or sample fixation [20].
Experimental Protocol: FF-OCT for Live-Cell Apoptosis Imaging
Objective: To monitor and characterize the morphological changes of apoptosis in live cells in a label-free manner using FF-OCT. Cell Line: HeLa cells (human cervical cancer cells). Reagents:
- Dulbecco’s Modified Eagle’s Medium (DMEM) for cell culture.
- Doxorubicin: Apoptosis-inducing agent. It intercalates into DNA and inhibits topoisomerase II, causing double-strand breaks and activating p53-mediated apoptosis [20].
- Ethanol (99%): Used as a positive control for inducing necrosis for comparative studies [20].
Procedure:
- Cell Culture and Preparation: Culture HeLa cells as a monolayer in DMEM under standard conditions (5% CO₂, 37°C). For experiments, plate cells onto imaging-compatible dishes or slides [20].
- Apoptosis Induction: To induce apoptosis, add doxorubicin to the culture medium at a final concentration of 5 μmol/L. For necrosis control, treat a separate group with 99% ethanol [20].
- FF-OCT Imaging:
- Utilize a custom-built time-domain FF-OCT system with a broadband halogen light source (e.g., center wavelength 650 nm) for sub-micrometer axial resolution [20].
- The system should be based on a Linnik-configured Michelson interferometer with identical high-numerical-aperture water-immersion objectives (e.g., 40x, NA: 0.8) in both reference and sample arms to achieve subcellular resolution [20].
- Initiate imaging immediately after drug administration.
- Acquire en face (x-y) cross-sectional images continuously at set intervals (e.g., every 20 minutes) for up to 180 minutes to monitor dynamic changes [20].
- Use a precision motorized stage to obtain z-stack images for 3D reconstruction.
- Image and Data Analysis:
- Reconstruct 3D surface topography by identifying the depth of maximum reflected intensity for each pixel to generate a 3D point cloud [20].
- Analyze changes in cell-substrate adhesion and cell boundary integrity using FF-OCT-based interference reflection microscopy (IRM)-like imaging [20].
- Quantify morphological parameters such as cell volume, surface roughness, and the presence of membrane blebs or spines over time.
The Scientist's Toolkit: Key Research Reagent Solutions
The following table details essential materials and reagents used in apoptosis morphology research.
Table 2: Essential Research Reagents and Materials for Apoptosis Morphology Studies
| Item Name | Function/Application | Specific Example |
|---|---|---|
| Doxorubicin | Chemotherapeutic agent used to induce intrinsic apoptosis via DNA damage and ROS generation [20]. | Final concentration of 5 μmol/L in cell culture medium [20]. |
| Ethanol | Chemical used to induce necrosis as a comparative control for cell death studies [20]. | 99% concentration applied to cell culture [20]. |
| Hoechst 33342 / DAPI / Acridine Orange (AO) | Fluorescent DNA-binding dyes for staining nuclei to visualize chromatin condensation and nuclear fragmentation via fluorescence microscopy [14]. | Used according to manufacturer's protocol for live (Hoechst, AO) or fixed (DAPI) cells. |
| Annexin V-FITC Apoptosis Detection Kit | Kit for flow cytometry or microscopy to detect phosphatidylserine (PS) exposure on the outer leaflet of the cell membrane, an early marker of apoptosis [54] [14]. | Often includes Propidium Iodide (PI) for dual staining to differentiate between viable, early apoptotic, and necrotic cells [54]. |
| Apoptosis Antibody Cocktails | Pre-mixed solutions of multiple antibodies for Western blot detection of key apoptosis markers (e.g., caspases, PARP, Bcl-2 family) in a single assay [57]. | Example: ab136812 cocktail targeting pro/p17-caspase-3, cleaved PARP1, and muscle actin [57]. |
| Caspase Antibodies | Primary antibodies for Western blot to detect the activation (cleavage) of initiator (caspase-8, -9) and executioner (caspase-3, -7) caspases [57]. | Critical for confirming apoptosis activation via biochemical methods. |
Apoptosis Signaling Pathways and Experimental Workflow
The following diagrams, generated using Graphviz DOT language, illustrate the core apoptosis signaling pathways and a generalized experimental workflow for morphological analysis.
Diagram 1: Core Apoptosis Signaling Pathways
Core Apoptosis Signaling Pathways Diagram. This illustrates the major extrinsic (death receptor) and intrinsic (mitochondrial) pathways that converge on the activation of executioner caspases, leading to the characteristic morphological changes of apoptosis [6] [17] [60].
Diagram 2: Experimental Workflow for Morphology Analysis
Experimental Workflow for Apoptosis Morphology Analysis. This flowchart outlines the key steps in a typical experiment, from inducing cell death and preparing samples to analyzing morphology through various techniques and interpreting the data [14] [57] [20].
The meticulous comparison of morphological features across apoptosis phases I, IIa, and IIb provides a critical foundation for understanding cellular fate in both physiological and pathological contexts. While traditional microscopy methods remain cornerstone techniques for identifying specific morphological hallmarks, advanced label-free technologies like FF-OCT are pushing the boundaries by enabling high-resolution, dynamic, and quantitative 3D analysis of living cells. The integration of these morphological assessments with biochemical techniques, such as Western blotting for caspase activation, offers a robust and comprehensive approach for researchers and drug developers to accurately detect and characterize apoptosis, thereby advancing our understanding of disease mechanisms and therapeutic efficacy.
Correlating Caspase Activation and PARP Cleavage with Morphological Progression
The morphological progression of apoptosis through its distinct phases—early (Phase I), middle (Phase IIa), and late (Phase IIb)—serves as the ultimate phenotypic manifestation of an intricate underlying molecular cascade. This technical guide delineates the precise correlation between two pivotal biochemical events, caspase activation and PARP cleavage, with the characteristic physical transformations of a dying cell. Framed within broader thesis research on apoptotic morphology, this synthesis of biochemical and morphological data provides researchers and drug development professionals with a structured framework to interpret experimental results and investigate cell death mechanisms. The controlled dismantling of a cell during apoptosis is not a chaotic process but a molecularly orchestrated event where specific biochemical hallmarks can be directly mapped to observable morphological stages [57].
The Apoptotic Program: Morphology and Molecular Drivers
Apoptosis, or programmed cell death, is a physiological process for eliminating damaged, infected, or superfluous cells in a controlled, non-inflammatory manner. This is in stark contrast to necrotic cell death, which involves cellular swelling and rupture, leading to inflammation [86] [57]. The apoptotic process can be triggered via two primary signaling pathways: the extrinsic pathway, initiated by extracellular death ligands binding to cell surface receptors, and the intrinsic pathway, initiated by intracellular stress signals such as DNA damage [57]. Both pathways converge on the activation of a specific family of proteases, the caspases, which execute the cell death program.
Morphological Progression of Apoptosis
The execution of apoptosis is characterized by a sequence of morphological changes, systematically categorized into three main phases [57]:
- Early Phase (Phase I): The cell begins to shrink, losing contact with its neighbors and surrounding extracellular matrix. The cytoplasm becomes denser due to water loss, and organelles remain largely intact. Early membrane changes occur, including the loss of microvilli and the translocation of phosphatidylserine from the inner to the outer leaflet of the plasma membrane.
- Middle Phase (Phase IIa): This phase is marked by profound nuclear alterations. Chromatin condenses and marginalizes, forming dense, compact masses against the nuclear envelope. The nucleus then undergoes fragmentation into discrete bodies.
- Late Phase (Phase IIb): The cell undergoes a process called "budding," where it forms multiple, membrane-bound apoptotic bodies containing nuclear debris, condensed cytoplasm, and intact organelles. The cytoskeleton is disassembled, and the cell surface exhibits pronounced blebbing. These apoptotic bodies are swiftly phagocytosed by neighboring cells or professional phagocytes, preventing an inflammatory response.
Core Molecular Events: Caspases and PARP
Caspase Activation: The Point of No Return
Caspases (cysteine-aspartic proteases) are a family of zymogens that, upon activation, cleave their target proteins after aspartic acid residues. They are the principal effectors of apoptosis, dismantling the cell by cleaving hundreds of cellular substrates [87]. Caspases are categorized as:
- Initiator Caspases (e.g., Caspase-8, -9): Activated in response to pro-apoptotic signals via proximity-induced autoprocessing in large multiprotein complexes. They then propagate the death signal by cleaving and activating downstream executioner caspases.
- Executioner Caspases (e.g., Caspase-3, -7): Once activated by initiator caspases, they systematically cleave a wide array of structural and functional proteins, leading to the characteristic morphological changes of apoptosis [57].
Table 1: Key Caspases in Apoptosis and Their Primary Roles
| Caspase | Role/Type | Activating Pathway | Key Functions/Substrates |
|---|---|---|---|
| Caspase-8 | Initiator | Extrinsic (Death Receptors) | Initiates the extrinsic pathway; cleaves and activates executioner caspases [57]. |
| Caspase-9 | Initiator | Intrinsic (Mitochondrial) | Activated by the Apaf-1 apoptosome; initiates the intrinsic pathway [57]. |
| Caspase-3 | Executioner | Both Extrinsic & Intrinsic | Primary executioner caspase; cleaves key substrates like PARP, DFF45/ICAD, leading to DNA fragmentation and cellular disassembly [57]. |
| Caspase-7 | Executioner | Both Extrinsic & Intrinsic | Works alongside Caspase-3 to execute the cell death program [57]. |
PARP Cleavage: A Biochemical Hallmark and Metabolic Switch
Poly (ADP-ribose) polymerase-1 (PARP-1) is a nuclear enzyme involved in DNA repair and genomic stability. Upon detecting DNA strand breaks, PARP-1 becomes activated and consumes large amounts of NAD+ to synthesize poly(ADP-ribose) chains on itself and other nuclear proteins. While this is beneficial for mild DNA damage, excessive PARP-1 activation can lead to severe ATP depletion, as the cell attempts to resynthesize NAD+, potentially causing necrotic cell death [86].
During apoptosis, executioner caspases, primarily caspase-3, cleave the 116-kDa PARP-1 protein at a specific DEVD motif (between Asp214 and Gly215). This cleavage event separates the PARP-1 DNA-binding domains from its catalytic domain, resulting in a 24-kDa fragment and an 89-kDa fragment (often detected as an 85-kDa band on Western blots due to further processing) and effectively inactivating the enzyme [86] [57]. This serves a critical biological function:
- Preventing Energy Depletion: By inactivating PARP-1, the apoptotic cell conserves cellular ATP (NAD+ and ATP pools are linked), which is required for the energy-dependent execution of the apoptotic program.
- Ensuring Apoptotic Fidelity: This cleavage acts as a "molecular switch" that ensures the cell dies by apoptosis rather than necrosis, thereby preventing the release of pro-inflammatory cellular contents [86].
- Facilitating Cellular Disassembly: Inactivation of PARP-1 may also prevent futile DNA repair efforts while the caspase-activated DNase is being set loose to fragment the genome.
Correlation: Molecular-Phenotypic Timeline
The activation of caspases and the subsequent cleavage of PARP are not isolated events but are tightly coupled to the morphological stages of apoptosis. The table below provides a phased correlation between these molecular and phenotypic events.
Table 2: Correlation of Caspase Activation and PARP Cleavage with Morphological Progression
| Apoptotic Phase | Key Molecular Events | Resulting Morphological Features |
|---|---|---|
| Early Phase (I) | Initiation of extrinsic/intrinsic pathways. Activation of initiator caspases (Caspase-8/-9). Initial activation of executioner caspases (Caspase-3/-7). | Cell shrinkage, cytoplasmic condensation, loss of specialized surface structures (e.g., microvilli). Exposure of "eat-me" signals like phosphatidylserine on the outer membrane [57]. |
| Middle Phase (IIa) | Peak activity of executioner caspases (Caspase-3/-7). Cleavage of key structural nuclear proteins (e.g., Lamin A). Cleavage of PARP. Activation of Caspase-Activated DNase (CAD). | Nuclear condensation (pyknosis) and nuclear fragmentation (karyorrhexis). Loss of nuclear integrity [57]. |
| Late Phase (IIb) | Widespread proteolysis by caspases of cytoskeletal and cytoplasmic proteins (e.g., Actin, Gelsolin). | Formation of apoptotic bodies. Extensive membrane blebbing. Final disintegration of the cell into multiple, membrane-bound vesicles ready for phagocytosis [57]. |
Experimental Detection and Analysis
Western Blot Protocol for Apoptosis Detection
Western blotting is a powerful and widely used technique for detecting specific proteins and their cleavage products during apoptosis, offering high specificity and the ability for quantification [57].
Sample Preparation:
- Cell Lysis: Lyse control and treated cells using a RIPA buffer or similar, supplemented with protease and phosphatase inhibitors to preserve protein integrity and phosphorylation states.
- Protein Quantification: Determine the protein concentration of each lysate using a standardized assay (e.g., BCA or Bradford assay). This is critical for loading equal amounts of protein across samples.
Gel Electrophoresis and Transfer:
- SDS-PAGE: Load an equal amount of total protein (e.g., 20-40 µg) per lane on a polyacrylamide gel. Separating gels between 10-15% are typically used to resolve apoptosis-related proteins (e.g., caspases ~17-55 kDa, PARP ~116 kDa & 89 kDa).
- Protein Transfer: Transfer the separated proteins from the gel onto a nitrocellulose or PVDF membrane.
Immunoblotting:
- Blocking: Incubate the membrane in a blocking solution (e.g., 5% non-fat milk or BSA in TBST) for 1 hour to prevent non-specific antibody binding.
- Primary Antibody Incubation: Incubate the membrane with primary antibodies specific for your target apoptotic markers. Key antibody targets include:
- Cleaved Caspase-3: Detects the active, large fragment (~17 kDa) of the executioner caspase.
- PARP: Detects both the full-length (116 kDa) and the cleaved (89 kDa) forms.
- Caspase-8, Caspase-9: To identify pathway-specific initiator caspase processing.
- Loading Control: Antibodies for housekeeping proteins like β-actin, GAPDH, or α-tubulin.
- Washing and Secondary Antibody Incubation: Wash the membrane and incubate with an appropriate horseradish peroxidase (HRP)-conjugated or fluorescently-labeled secondary antibody.
Detection and Visualization: Develop the blot using chemiluminescent, fluorescent, or colorimetric detection methods. Capture the image using a digital imager.
Interpreting Western Blot Results
- Caspase Activation: Is indicated by the disappearance of the pro-caspase band (e.g., 32-35 kDa for pro-caspase-3) and/or the appearance of its cleaved, active fragments (e.g., 17 kDa and 12 kDa for caspase-3) [57].
- PARP Cleavage: Is a definitive marker of apoptosis. A successful apoptotic induction is shown by the decrease in the full-length PARP band (116 kDa) and the concomitant increase in the cleaved PARP fragment (89 kDa) [57].
- Quantification:
- Use densitometry software (e.g., ImageJ) to measure the band intensities.
- Calculate the ratio of cleaved protein to total protein (e.g., Cleaved Caspase-3 / Total Caspase-3) or cleaved to full-length protein (e.g., Cleaved PARP / Full-length PARP).
- Normalize these ratios to a loading control (e.g., β-actin) to account for any variations in sample loading and transfer efficiency. This allows for comparative analysis across different experimental conditions.
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Studying Caspase Activation and PARP Cleavage
| Reagent / Assay | Function / Application |
|---|---|
| Annexin V-FITC/PI Staining | Flow cytometry assay to detect early apoptosis (phosphatidylserine exposure) and late apoptosis/necrosis (membrane permeability) [88]. |
| Caspase Inhibitor (e.g., zVAD-fmk) | A pan-caspase inhibitor used to confirm the caspase-dependent nature of cell death. Note: It can potentiate necrosis in some TNF-induced death models [86]. |
| PARP Inhibitor (e.g., 3-AB) | Used to investigate the role of PARP activity in cell death and to mimic the energy-conserving effects of its cleavage [86]. |
| Antibody Cocktails (e.g., ab136812) | Pre-mixed antibodies targeting multiple apoptosis markers (e.g., pro/p17-caspase-3, cleaved PARP, actin). They streamline workflows, enhance detection, and improve reproducibility [57]. |
| Carbon Nanoparticles (CDots) | Novel fluorescent nanoparticles reported to show increased accumulation and altered distribution in apoptotic cells, potentially useful for imaging and flow cytometry [89]. |
Signaling Pathway and Experimental Workflow Visualization
Apoptotic Signaling Cascade and PARP Cleavage
Experimental Workflow for Correlation Analysis
The precise correlation between caspase-mediated PARP cleavage and the systematic morphological dismantling of a cell provides a robust framework for understanding and investigating apoptotic cell death. The molecular events—initiator caspase activation, executioner caspase amplification, and the decisive cleavage of PARP—serve as definitive biochemical markers that can be mapped directly onto the phases of morphological progression. This integrated understanding is fundamental for research in cancer biology, neurodegenerative diseases, and drug development, where modulating apoptosis is a key therapeutic strategy. Western blot analysis, complemented by morphological techniques, remains a cornerstone for experimentally validating this correlation and advancing our knowledge of cell death biology.
The comprehensive analysis of cell death, particularly apoptosis, is a cornerstone of experimental oncology and therapeutic development. Apoptosis is a tightly regulated process characterized by distinct morphological phases: Phase I (cell shrinkage, dense cytoplasm), Phase IIa (chromatin condensation, nuclear fragmentation), and Phase IIb (membrane blebbing, apoptotic body formation) [14] [17]. Relying on morphological assessment alone, however, provides limited insight into the underlying molecular mechanisms driving cell death. Conversely, molecular techniques alone may miss crucial contextual information about cellular state and death progression. The integration of both approaches provides a powerful framework for validating experimental findings and obtaining a holistic understanding of therapeutic action.
This technical guide examines the strategic application of combined morphological and molecular analyses through a detailed case study in breast cancer research, providing methodologies and resources to implement this approach effectively.
Integrated Analysis Case Study: Thymoquinone and Methotrexate in Breast Cancer
A 2025 study on MCF-7 estrogen receptor-positive breast cancer cells provides an exemplary model of integrated morphological and molecular analysis to investigate the synergistic effects of thymoquinone (TQ) and methotrexate (MTX) [90].
The study employed a sequential workflow to first identify phenotypic effects and then investigate the molecular mechanisms responsible.
Table 1: Summary of Key Quantitative Findings from Combination Treatment
| Analysis Parameter | TQ Alone (100 µM) | MTX Alone (10 µM) | Combination (100 µM TQ + 10 µM MTX) |
|---|---|---|---|
| Cell Viability (24h) | ~40% | ~50% | <20% |
| Total Apoptosis | 37.4% | 68.3% | 83.6% |
| ROS Increase | ~3-fold | ~4-fold | ~6-fold |
| Bax/Bcl-2 Ratio | Significantly increased | Significantly increased | Most pronounced increase |
| Caspase-3 Activation | Moderate | Moderate | Markedly enhanced |
| Cell Cycle Arrest | Moderate G2/M phase arrest | Moderate G2/M phase arrest | Pronounced G2/M phase arrest |
Source: Data compiled from [90]
Morphological Assessment Techniques
The initial investigation focused on characterizing the morphological hallmarks of apoptosis:
- Phase I and IIa Analysis: Cells were stained with Hoechst 33342 or DAPI for fluorescence microscopy to visualize nuclear condensation (pyknosis) and chromatin margination, key features of early and middle apoptotic stages [14].
- Phase IIb Analysis: Membrane blebbing and apoptotic body formation were assessed using time-lapse quantitative phase imaging (QPI), which enabled label-free observation of subtle changes in cell mass distribution and morphology [91].
- Apoptosis Quantification: Annexin V/PI flow cytometry was employed to distinguish intact cells (Annexin V-/PI-), early apoptotic cells (Annexin V+/PI-, exhibiting Phase I/IIa morphology), and late apoptotic/necrotic cells (Annexin V+/PI+) [90] [14].
Molecular Analysis Techniques
Following morphological confirmation of apoptosis, researchers deployed molecular techniques to decipher the underlying signaling pathways:
- Caspase Activation: Western blot analysis detected cleaved fragments of executioner caspases-3 and -7, providing definitive evidence of apoptotic protease cascade activation [90] [57].
- Gene Expression Regulation: qRT-PCR analysis demonstrated combination treatment upregulated pro-apoptotic Bax, downregulated anti-apoptotic Bcl-2, and suppressed metastasis-related genes (NF-κB, MMP-2, MMP-9) [90].
- Oxidative Stress Measurement: Intracellular ROS generation was quantified fluorometrically using DCFH-DA assay, revealing an approximately six-fold increase with combination therapy [90].
- Mitochondrial Pathway Assessment: Spectrophotometric analysis of antioxidant enzymes (SOD, CAT) showed significant suppression of activity, indicating profound oxidative stress induction [90].
Diagram 1: Integrated molecular and morphological pathway of TQ+MTX-induced apoptosis. The combination therapy triggers molecular events (red) culminating in apoptosis, which manifests through characteristic morphological phases (blue).
Core Methodologies for Combined Analysis
Morphological Characterization Techniques
Light Microscopy with Histochemical Staining
- Protocol: Culture cells on chamber slides, treat with experimental compounds, fix with 4% PFA, and stain with Hematoxylin and Eosin (H&E) or Giemsa [14].
- Detection Capability: Identifies late-stage apoptosis (Phase IIb) features including cell shrinkage, nuclear fragmentation, and apoptotic body formation [14].
- Advantages/Limitations: Simple and intuitive, but lacks sensitivity for early apoptosis detection and requires complementary molecular validation [14].
Fluorescence Microscopy with Nuclear Stains
- Protocol: Stain live or fixed cells with Hoechst 33342, DAPI, or acridine orange (1-5 µg/mL for 10-30 minutes), then visualize using fluorescence or confocal microscopy [14] [17].
- Detection Capability: Reveals nuclear morphology changes characteristic of Phase IIa apoptosis, including chromatin condensation and nuclear fragmentation [14].
- Advantages/Limitations: Provides superior nuclear detail but offers limited information about cytoplasmic events [14].
Quantitative Phase Imaging (QPI)
- Protocol: Perform time-lapse imaging of unlabeled cells using specialized QPI systems, then analyze parameters like cell density and dynamic morphology changes [91].
- Detection Capability: Quantifies subtle mass distribution changes throughout all apoptosis phases, distinguishing apoptosis from necrosis with ~76% accuracy [91].
- Advantages/Limitations: Label-free, continuous monitoring but requires specialized instrumentation [91].
Transmission Electron Microscopy
- Protocol: Fix cells with glutaraldehyde, post-fix with osmium tetroxide, dehydrate, embed in resin, section, and stain with uranyl acetate-lead citrate [14] [17].
- Detection Capability: Visualizes ultrastructural features of all apoptosis phases, including organelle changes, cavitation (Phase I), chromatin margination (Phase IIa), and apoptotic bodies (Phase IIb) [14].
- Advantages/Limitations: Provides highest resolution morphological details but is resource-intensive and low-throughput [14].
Molecular Characterization Techniques
Western Blot Analysis for Apoptosis Markers
- Sample Preparation: Lyse cells in RIPA buffer with protease/phosphatase inhibitors. Quantify protein concentration using BCA assay [57].
- Electrophoresis and Transfer: Separate 20-50 µg protein via SDS-PAGE (4-20% gradient gels), then transfer to PVDF membranes [57].
- Antibody Probing: Block with 5% BSA, incubate with primary antibodies (cleaved caspase-3, PARP, Bax, Bcl-2) overnight at 4°C, then with HRP-conjugated secondary antibodies for 1 hour [57].
- Detection and Analysis: Use chemiluminescent substrate and image with CCD system. Normalize band intensity to housekeeping proteins (β-actin, GAPDH) [57].
Flow Cytometry for Apoptosis Quantification
- Annexin V/PI Staining: Harvest cells, wash in PBS, resuspend in binding buffer with FITC-Annexin V and PI (1 µg/mL), incubate 15 minutes in dark, and analyze within 1 hour [90] [14].
- Data Interpretation: Viable cells (Annexin V-/PI-); Early apoptotic (Annexin V+/PI-, Phase I/IIa); Late apoptotic (Annexin V+/PI+, Phase IIb); Necrotic (Annexin V-/PI+) [14].
Gene Expression Analysis
- qRT-PCR Protocol: Extract RNA, reverse transcribe to cDNA, then amplify with gene-specific primers (Bax, Bcl-2, caspase family, survivin) using SYBR Green chemistry [90] [92].
- Data Analysis: Calculate fold changes using ΔΔCt method, normalizing to housekeeping genes (GAPDH, β-actin) [90].
Table 2: Methodological Approaches for Apoptosis Phase Analysis
| Apoptosis Phase | Key Morphological Features | Primary Morphological Techniques | Confirmatory Molecular Assays |
|---|---|---|---|
| Phase I (Early) | Cell shrinkage, dense cytoplasm, loss of microvilli | TEM, QPI | Mitochondrial membrane potential assay, Annexin V staining (without PI) |
| Phase IIa (Middle) | Chromatin condensation, nuclear pyknosis, ribosomal dissociation | Fluorescence microscopy (Hoechst/DAPI), TEM | Caspase-3/7 activation assays, Western blot for cleaved caspases |
| Phase IIb (Late) | Membrane blebbing, apoptotic body formation, organelle packaging | Light microscopy (H&E), QPI, TEM | DNA fragmentation assay (TUNEL), PI staining, PARP cleavage detection |
Source: Data compiled from [14] [17]
Table 3: Research Reagent Solutions for Combined Apoptosis Analysis
| Reagent/Category | Specific Examples | Research Application |
|---|---|---|
| Viability/Cytotoxicity Assays | MTT, MTS, WST-1 | Preliminary screening of treatment efficacy and IC50 determination [90] |
| Apoptosis Detection Kits | Annexin V-FITC/PI apoptosis detection kits | Flow cytometry-based quantification of apoptotic populations [90] [14] |
| Caspase Activity Assays | CellEvent Caspase-3/7 Green Detection Reagent, fluorogenic substrates | Detection of initiator and executioner caspase activation [91] [18] |
| Antibody Cocktails | Pro/p17-caspase-3, cleaved PARP1, muscle actin (ab136812) | Simultaneous detection of multiple apoptosis markers by Western blot [57] |
| Nuclear Stains | Hoechst 33342, DAPI, Propidium Iodide (PI) | Morphological assessment of nuclear changes during apoptosis [14] |
| ROS Detection Probes | DCFH-DA, Dihydroethidium | Measurement of reactive oxygen species generation [90] |
| Mitochondrial Probes | JC-1, TMRM, MitoTracker | Assessment of mitochondrial membrane potential and mass [14] |
| qPCR Assays | Primer sets for Bax, Bcl-2, caspases, survivin, MMPs | Gene expression analysis of apoptosis regulators [90] [92] |
Source: Data compiled from multiple sources [90] [91] [14]
Diagram 2: Experimental workflow for combined morphological and molecular analysis. The parallel approaches converge for data integration, providing complementary evidence for mechanistic conclusions.
The integration of morphological and molecular analyses creates a powerful framework for comprehensive apoptosis assessment in cancer research. The case study of thymoquinone and methotrexate in MCF-7 cells demonstrates how this combined approach can elucidate complex therapeutic interactions, revealing synergistic induction of apoptosis through oxidative stress, caspase activation, and cell cycle arrest while characterizing the morphological progression through definitive apoptotic stages.
This methodology overcomes the limitations of single-technique approaches, where morphological assessment alone might lack mechanistic insight, and molecular data alone might miss crucial contextual information about cellular state. The strategic combination of these techniques provides robust, multi-dimensional validation of experimental findings, accelerating therapeutic development and enhancing our understanding of cancer biology.
The Role of Morphological Assessment in Validating Novel Apoptosis-Targeting Therapies
Apoptosis, or programmed cell death, is a fundamental process crucial for maintaining tissue homeostasis, eliminating damaged or infected cells, and ensuring proper embryonic development [17] [6]. Its significance is profoundly amplified in the context of cancer therapy, where the primary goal of many treatment modalities is to induce apoptotic cell death in malignant cells [93]. The morphological features of apoptosis serve as the definitive hallmark of this form of cell death, providing a visible manifestation of the underlying biochemical cascade [14]. While the molecular mechanisms of apoptosis are well-delineated, encompassing both intrinsic and extrinsic pathways, the ultimate validation of a successful pro-apoptotic therapy lies in the direct observation of characteristic cellular changes [17] [6]. This guide delves into the critical role of morphological assessment in validating novel apoptosis-targeting therapies, framing its discussion within the broader thesis of apoptosis phase research (Phases I, IIa, and IIb) and providing researchers with advanced methodologies for robust experimental analysis.
Morphological Hallmarks of Apoptosis Phases
The execution of apoptosis follows a sequential pattern, classically divided into three phases based on distinct morphological alterations in the nucleus and cytoplasm. These phases provide a critical framework for identifying and quantifying apoptotic cells in response to therapeutic agents.
- Phase I: The initial phase is characterized by cell shrinkage and a reduction in cell volume. The cytoplasm becomes denser due to dehydration, and eosinophilia increases. Notably, specialized cell-surface structures like microvilli begin to disappear, and the cell detaches from its neighboring cells and the extracellular matrix [14].
- Phase IIa: This stage involves dramatic changes in the nucleus, known as chromatin condensation. The chromatin aggregates into dense, featureless masses in a process called pyknosis, or becomes marginalized along the inner nuclear membrane. Subsequently, the nucleus breaks apart into discrete fragments, a stage termed nuclear fragmentation [14].
- Phase IIb: The final phase involves the packaging of the cell into apoptotic bodies. The cell membrane undergoes intricate sprouting and blebbing, eventually pinching off to form membrane-bound vesicles. These apoptotic bodies contain well-preserved organelles and nuclear debris. A key feature is the maintenance of plasma membrane integrity, which prevents the release of cellular contents and avoids a pro-inflammatory response [14]. The apoptotic bodies are swiftly recognized and phagocytosed by nearby macrophages and other cells, leaving no trace [14].
Table 1: Morphological Characteristics of Apoptosis Phases
| Apoptosis Phase | Nuclear Changes | Cytoplasmic Changes | Cellular Outcome |
|---|---|---|---|
| Phase I | Minor chromatin condensation | Cell shrinkage, increased eosinophilia, loss of microvilli, dissociation from neighbors | Preparation for dissolution |
| Phase IIa | Pronounced chromatin condensation (pyknosis), nuclear fragmentation (karyorrhexis) | Continued condensation, dilation of endoplasmic reticulum | Nuclear disintegration |
| Phase IIb | Nuclear fragments packaged into apoptotic bodies | Membrane blebbing and sprouting, formation of apoptotic bodies containing organelles | Formation of apoptotic bodies for phagocytosis |
Molecular Mechanisms and Signaling Pathways
The morphological changes observed during apoptosis are the direct result of a tightly regulated molecular cascade. Apoptosis can be initiated via two primary pathways: the extrinsic (death receptor) pathway and the intrinsic (mitochondrial) pathway, which converge on a common execution phase.
The Extrinsic Pathway: This pathway is triggered by the binding of extracellular death ligands (e.g., FasL, TRAIL, TNF-α) to their corresponding death receptors on the cell surface [6]. This binding recruits adapter proteins like FADD to form the Death-Inducing Signaling Complex (DISC), which activates initiator caspases, primarily caspase-8 and caspase-10 [17] [93]. Active caspase-8 can directly cleave and activate downstream effector caspases.
The Intrinsic Pathway: Initiated by intracellular stressors such as DNA damage, oxidative stress, or growth factor withdrawal, this pathway is regulated by the BCL-2 protein family [6] [93]. Pro-apoptotic proteins like BAX and BAK oligomerize and permeabilize the mitochondrial outer membrane (MOMP), leading to the release of cytochrome c into the cytosol [17] [93]. Cytochrome c binds to APAF-1, forming the "apoptosome," which activates the initiator caspase-9 [93].
Both pathways converge on the execution phase, where initiator caspases (caspase-8, -9, -10) activate effector caspases (caspase-3, -6, -7) [17]. Caspase-3 is a key executioner caspase that cleaves a multitude of cellular substrates, including structural proteins and DNA repair enzymes, leading to the characteristic biochemical and morphological hallmarks of apoptosis, such as DNA fragmentation and cell shrinkage [17] [14]. The cleavage of PARP is another biomarker of apoptosis, preventing DNA repair and facilitating cellular disassembly [6].
Advanced Morphological Assessment Techniques
Validating apoptosis-inducing therapies requires a multifaceted approach that combines qualitative morphological observation with quantitative assays.
Microscopy-Based Techniques
- Light Microscopy: After staining with hematoxylin and eosin (H&E), Giemsa, or Wright's stain, apoptotic cells can be identified by cell shrinkage, nuclear condensation, and the presence of apoptotic bodies. This method is simple and intuitive but is mainly suitable for observing Phase IIb apoptosis, as early phases and small-scale apoptosis can be easily missed [14].
- Transmission Electron Microscopy (TEM): TEM is the gold standard for visualizing the ultrastructural features of apoptosis [14]. It can reveal early changes like cavitation (Phase I), highly condensed and marginalized chromatin (Phase IIa), and the detailed structure of apoptotic bodies (Phase IIb). Its major advantage is the unparalleled resolution of intracellular structures.
- Fluorescence/Confocal Microscopy: Using DNA-binding fluorescent dyes like Hoechst 33342, DAPI, or Acridine Orange, researchers can observe nuclear morphology in detail [14]. Apoptotic nuclei exhibit intense, condensed, or fragmented staining compared to the diffuse staining of viable cells. This technique is particularly powerful for identifying Phase IIa and IIb apoptosis and can be combined with immunohistochemical staining.
Quantitative and High-Throughput Methods
- DNA Gel Electrophoresis: Apoptotic cells exhibit internucleosomal DNA cleavage, yielding a characteristic "ladder" pattern of DNA fragments (180-200 bp and multiples) upon gel electrophoresis. This method is a classic biochemical confirmation but is only semi-quantitative and best for detecting middle to late-stage apoptosis in large cell populations [14].
- TUNEL Assay: The TUNEL (TdT dUTP Nick-End Labeling) assay enzymatically labels the 3'-OH ends of DNA fragments, providing a relatively sensitive and specific way to detect and quantify apoptotic cells in situ. It is highly suitable for detecting late-stage apoptosis, though false positives can occur, necessitating proper controls [14].
- Imaging Flow Cytometry: This technology combines the high-throughput statistical power of flow cytometry with the visual confirmation of microscopy [94]. It allows for the acquisition of brightfield, darkfield, and multiple fluorescent images of thousands of cells per second. This enables not only the quantification of apoptosis based on fluorescence (e.g., with Annexin V or caspase substrates) but also the concurrent morphological verification of each event, drastically improving the accuracy of analysis in heterogeneous samples [94].
Table 2: Comparison of Key Apoptosis Detection Methodologies
| Method | Principle | Key Readout | Advantages | Limitations | Suitable Apoptosis Phase |
|---|---|---|---|---|---|
| Light Microscopy | Morphological staining | Cell shrinkage, nuclear condensation, apoptotic bodies | Simple, intuitive, storable specimens | Misses early/small-scale apoptosis | Phase IIb |
| Electron Microscopy | Ultrastructure visualization | Chromatin margination, organelle integrity, apoptotic bodies | Unparalleled resolution, reveals early phases | Technically demanding, low throughput | Phases I, IIa, IIb |
| Fluorescence Microscopy | Nuclear dye fluorescence | Condensed/fragmented nuclei | Detailed nuclear assessment, combinable | Subjective quantification | Phases IIa, IIb |
| DNA Gel Electrophoresis | DNA fragmentation | DNA "ladder" pattern | Classic biochemical confirmation | Semi-quantitative, no cell localization | Middle to Late Stage |
| TUNEL Assay | Labeling of DNA breaks | Labeled 3'-OH ends in situ | Sensitive, specific, allows quantification | Potential for false positives | Late Stage |
| Imaging Flow Cytometry | Cell imaging in flow | Morphology + fluorescence of single cells | High-throughput, multiparametric, quantitative | Expensive instrumentation, complex data | All Phases |
Experimental Workflow for Therapy Validation
A robust workflow for validating an apoptosis-targeting therapy integrates multiple techniques to provide comprehensive evidence, from initial screening to mechanistic insight.
Detailed Methodologies for Key Experiments
Protocol 1: Morphological Analysis via Fluorescence Microscopy
- Cell Seeding and Treatment: Seed cells onto glass coverslips placed in a culture dish. After adherence, treat with the investigational therapeutic agent.
- Staining: At designated time points, fix cells with 4% paraformaldehyde for 15 minutes. Permeabilize with 0.1% Triton X-100 for 10 minutes. Incubate with a nuclear stain (e.g., Hoechst 33342 at 1 µg/mL or DAPI) for 10-15 minutes in the dark.
- Mounting and Imaging: Mount coverslips onto glass slides using an anti-fade mounting medium. Visualize using a fluorescence microscope with appropriate filters. Apoptotic cells are identified by intensely stained, condensed, or fragmented nuclei.
Protocol 2: High-Throughput Quantification via Imaging Flow Cytometry
- Sample Preparation: Harvest treated and control cells. For early apoptosis detection, stain with Annexin V-FITC in a calcium-containing binding buffer. To discriminate late apoptosis/necrosis, include Propidium Iodide (PI). For caspase activation, use a cell-permeable fluorescent caspase inhibitor (e.g., FLICA).
- Instrument Acquisition: Acquire data on an imaging flow cytometer (e.g., ImageStream). Collect brightfield, darkfield (side scatter), and fluorescence images for at least 10,000 cells per sample. Use a low flow rate for higher image clarity.
- Data Analysis: Using instrument software, first gate on single, intact cells based on brightfield area and aspect ratio. Then, identify Annexin V-positive and/or PI-positive populations. Critically, review the images of gated populations to confirm morphological features of apoptosis (cell shrinkage, membrane blebbing, nuclear condensation) and exclude artifacts.
Protocol 3: Biochemical Confirmation via Western Blot
- Protein Extraction: Lyse cells in RIPA buffer supplemented with protease and phosphatase inhibitors.
- Electrophoresis and Transfer: Separate equal amounts of protein by SDS-PAGE and transfer to a PVDF membrane.
- Immunoblotting: Probe the membrane with primary antibodies against key apoptotic biomarkers: Cleaved Caspase-3 (the active executioner caspase), Cleaved PARP (a marker of irreversible apoptotic commitment), and a loading control (e.g., GAPDH or β-Actin). The appearance of cleaved bands is a definitive molecular correlate of apoptosis induction.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Reagents for Apoptosis Morphology Research
| Reagent / Assay | Function / Target | Key Application in Apoptosis Detection |
|---|---|---|
| Hoechst 33342 / DAPI | DNA-binding fluorescent dyes | Visualization of nuclear condensation and fragmentation (Phases IIa, IIb) via fluorescence microscopy. |
| Annexin V-FITC/PI Assay | Binds phosphatidylserine (PS) / DNA intercalator | Flow cytometry or imaging flow cytometry to detect PS externalization (early apoptosis) and loss of membrane integrity (late apoptosis/necrosis). |
| FLICA (Fluorescent-Labeled Inhibitor of Caspases) | Irreversibly binds active caspases | Direct detection and quantification of caspase activation (e.g., caspase-3) in live cells by flow cytometry. |
| TUNEL Assay Kit | Labels DNA strand breaks | In situ detection and quantification of late-stage apoptotic cells with DNA fragmentation in cell cultures or tissue sections. |
| Antibody: Anti-Cleaved Caspase-3 | Detects activated caspase-3 | Gold-standard immunohistochemical or Western Blot confirmation of executioner caspase activation. |
| Antibody: Anti-Cleaved PARP | Detects inactivated PARP | Western Blot biomarker for irreversible apoptotic commitment and DNA damage response. |
| Primary Antibody Panels (CD45, CD3, etc.) | Cell surface and intracellular markers | Phenotypic identification of specific cell types (e.g., immune cells) within a heterogeneous population during co-culture or ex vivo analysis via imaging flow cytometry [94]. |
Morphological assessment remains an indispensable pillar in the validation of novel apoptosis-targeting therapies. While molecular and biochemical assays provide critical data on pathway activation, the observation of the characteristic morphological sequence—cell shrinkage, chromatin condensation, and apoptotic body formation—provides unequivocal proof of a cell's commitment to apoptotic death. The integration of classical microscopy with modern high-throughput technologies like imaging flow cytometry offers a powerful paradigm, enabling researchers to not only quantify apoptotic responses with statistical rigor but also to visually confirm the phenotype of every measured event. As cancer therapies grow more complex, aiming to overcome resistance by targeting specific nodes in the apoptotic machinery, a rigorous, morphology-centered validation strategy will be paramount for accurately assessing therapeutic efficacy and advancing successful candidates toward clinical application.
Conclusion
The precise identification of morphological features across apoptosis Phase I, IIa, and IIb remains a cornerstone of cell death research. A thorough understanding of these physical changes—from initial cell shrinkage to the formation of apoptotic bodies—enables accurate interpretation of experimental results and clinical biopsies. While morphological analysis provides intuitive and critical evidence, its power is maximized when integrated with biochemical and molecular techniques like caspase detection and TUNEL assays. This multi-modal approach is essential for advancing drug discovery, particularly in oncology and neurodegenerative diseases, where modulating apoptosis is a key therapeutic strategy. Future research will continue to refine these techniques and further elucidate the intricate relationship between cellular structure and death signaling pathways.
References
- 1. Morphologic and biochemical hallmarks of apoptosis [https://pubmed.ncbi.nlm.nih.gov/10728374/]
- 2. Apoptosis [https://en.wikipedia.org/wiki/Apoptosis]
- 3. Cell Death - an overview [https://www.sciencedirect.com/topics/materials-science/cell-death]
- 4. Targeting the Met-RIPK1 signaling axis to enforce ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12537949/]
- 5. ON/OFF and Beyond - A Boolean Model of Apoptosis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2781112/]
- 6. Insights on the crosstalk among different cell death ... [https://www.nature.com/articles/s41420-025-02328-9]
- 7. Selective Control of the Apoptosis Signaling Network in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1890306/]
- 8. Apoptosis - StatPearls - NCBI Bookshelf [https://www.ncbi.nlm.nih.gov/books/NBK499821/]
- 9. 50 years on and still very much alive: 'Apoptosis [https://www.nature.com/articles/s41416-022-02020-0]
- 10. Apoptosis Overview | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/life-science/antibodies/antibodies-learning-center/antibodies-resource-library/antibody-methods/apoptosis-overview.html]
- 11. Apoptosis pathway-targeted drugs—from the bench to ... [https://www.sciencedirect.com/science/article/abs/pii/S0304419X04000617]
- 12. Biomarkers of apoptosis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2538762/]
- 13. Apoptosis: A Comprehensive Overview of Signaling Pathways ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11592877/]
- 14. Apoptosis detection: a purpose-dependent approach ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8208110/]
- 15. Detection and Quantification of Nuclear Morphology ... [https://ar.iiarjournals.org/content/37/5/2239]
- 16. Chromatin Collapse during Caspase-dependent Apoptotic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3610992/]
- 17. Research progress on morphology and mechanism of ... [https://www.nature.com/articles/s41419-024-06712-8]
- 18. A quantitative real-time approach for discriminating ... [https://www.nature.com/articles/cddiscovery2016101]
- 19. Apoptosis: A Comprehensive Overview of Signaling ... [https://www.mdpi.com/2073-4409/13/22/1838]
- 20. High-Resolution Imaging of Morphological Changes ... [https://www.mdpi.com/2079-6374/15/8/522]
- 21. Deciphering DED assembly mechanisms in FADD ... [https://www.nature.com/articles/s41467-024-47990-2]
- 22. Reactive Oxygen Species Across Death Pathways [https://pmc.ncbi.nlm.nih.gov/articles/PMC12564638/]
- 23. Apoptosis, necroptosis, and pyroptosis as alternative cell ... [https://www.sciencedirect.com/science/article/pii/S0304419X23001737]
- 24. Apoptotic cell clearance: basic biology and therapeutic potential [https://pmc.ncbi.nlm.nih.gov/articles/PMC4040260/]
- 25. Phagocytosis of Apoptotic Cells in Resolution of Inflammation [https://pmc.ncbi.nlm.nih.gov/articles/PMC7137555/]
- 26. Phagocyte receptors for apoptotic cells: recognition, uptake ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC200959/]
- 27. Tailoring of apoptotic bodies for diagnostic and therapeutic ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-024-05451-w]
- 28. Regulation of Apoptotic Cell Clearance During Resolution ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2019.00891/full]
- 29. Assessment of phagocytic activity in live macrophages ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6994062/]
- 30. Morphological assessment of apoptosis [https://www.sciencedirect.com/science/article/abs/pii/S1046202307002150]
- 31. Guidelines for Regulated Cell Death Assays: A Systematic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7970050/]
- 32. Histochemical detection and comparison of apoptotic cells ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4976550/]
- 33. Ultrastructural and Morphological Effects in T ... [https://www.mdpi.com/1420-3049/27/13/4136]
- 34. 4.5. Giemsa Staining for the Detection of Morphological ... [https://bio-protocol.org/exchange/minidetail?id=6090221&type=30]
- 35. White blood cell detection, classification and analysis using ... [https://www.nature.com/articles/s41598-022-21250-z]
- 36. AI-based Apoptosis Cell Classification Using Phase ... [https://ar.iiarjournals.org/content/44/3/935]
- 37. Identifying and Quantifying Apoptosis: Navigating ... [https://www.nature.com/articles/3880776]
- 38. Detection of apoptosis : A review of conventional and novel... [https://pubs.rsc.org/en/content/articlehtml/2010/ay/c0ay00247j]
- 39. – Pathologia Apoptosis [https://pathologia.ed.ac.uk/topic/apoptosis/]
- 40. Uncovering the bequeathing potential of apoptotic ... [https://stemcellres.biomedcentral.com/articles/10.1186/s13287-025-04370-x]
- 41. Pharmacological inhibition of apoptosis, necroptosis, and ... [https://www.nature.com/articles/s41598-025-17275-9]
- 42. Researchers identify new molecular switch involved in ... [https://www.news-medical.net/news/20251103/Researchers-identify-new-molecular-switch-involved-in-programmed-cell-death.aspx]
- 43. Hoechst 33342 Protocol for Imaging [https://www.thermofisher.com/us/en/home/references/protocols/cell-and-tissue-analysis/protocols/hoechst-33342-imaging-protocol.html]
- 44. Cell Staining with Hoechst or DAPI Nuclear Stains [https://biotium.com/tech-tips-protocols/protocol-staining-cells-with-hoechst-or-dapi-nuclear-stains/]
- 45. Density imaging of heterochromatin in live cells using ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5687035/]
- 46. Confocal Microscopy: Principles and Modern Practices - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6961134/]
- 47. Resolution and Contrast in Confocal Microscopy [https://evidentscientific.com/en/microscope-resource/knowledge-hub/techniques/confocal/resolutionintro]
- 48. The molecular mechanisms and therapeutic implications of ... [https://link.springer.com/article/10.1007/s10495-025-02157-2]
- 49. Imaging Flow Cytometry as a Molecular Biology Tool [https://pmc.ncbi.nlm.nih.gov/articles/PMC12524540/]
- 50. Imaging flow cytometry: from high - resolution ... [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2025.1580749/full]
- 51. Comparing Flow Cytometry with Imaging Cytometry [https://www.labx.com/resources/comparing-flow-cytometry-with-imaging-cytometry/5523]
- 52. An in vivo microscopy dataset for the characterization of ... [https://www.nature.com/articles/s41597-025-04632-6]
- 53. Transformer-based spatial–temporal detection of apoptotic ... [https://elifesciences.org/articles/90502]
- 54. Apoptosis Assay Market Size Report, 2025 – 2034 [https://www.gminsights.com/industry-analysis/apoptosis-assay-market]
- 55. Apoptosis Testing Market Size, Demand & Forecast 2025- ... [https://www.futuremarketinsights.com/reports/apoptosis-testing-market]
- 56. Apoptosis Kit 2025 Trends and Forecasts 2033 [https://www.archivemarketresearch.com/reports/apoptosis-kit-136979]
- 57. Apoptosis western blot guide [https://www.abcam.com/en-us/knowledge-center/western-blot/western-blot-analysis-of-apoptosis]
- 58. The Quantitative-Phase Dynamics of Apoptosis and Lytic ... [https://www.nature.com/articles/s41598-020-58474-w]
- 59. Characterization and identification of cell death dynamics by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9047449/]
- 60. Comprehensive landscape of cell death mechanisms [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1611055/full]
- 61. Mechanisms of Apoptosis and Autophagy in Cell Life and Death [https://pmc.ncbi.nlm.nih.gov/articles/PMC8327148/]
- 62. Dual Topoisomerase I/II Inhibition-Induced Apoptosis and ... [https://www.mdpi.com/1420-3049/27/22/7993]
- 63. Apoptosis and Beyond: Cytometry in Studies of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3263828/]
- 64. Morphologic criteria and detection of apoptosis [https://pubmed.ncbi.nlm.nih.gov/10412642/]
- 65. Apoptosis: A Review of Programmed Cell Death - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2117903/]
- 66. Mechanisms of Cell Death: Apoptosis | Intrinsic & Extrinsic ... [https://blog.cellsignal.com/mechanisms-of-cell-death-apoptosis]
- 67. In Vitro Cell Death Determination for Drug Discovery [https://pmc.ncbi.nlm.nih.gov/articles/PMC5392044/]
- 68. Phenotypic high-throughput screening platform identifies ... [https://www.nature.com/articles/s41420-020-0240-0]
- 69. Flow cytometry-based apoptosis detection - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3863590/]
- 70. A new way of measuring apoptosis by absolute quantitation of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3424536/]
- 71. Apoptosis and the Response to Anti-Cancer Drugs [https://www.mayo.edu/research/labs/anticancer-drug-action/research-projects/apoptosis-response-anticancer-drugs]
- 72. Apoptotic cell death in disease—Current understanding of ... [https://www.nature.com/articles/s41418-023-01153-w]
- 73. Flow cytometry-based quantitative analysis of cellular ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11260931/]
- 74. Annexin V Staining Protocol for Flow Cytometry [https://www.thermofisher.com/us/en/home/references/protocols/cell-and-tissue-analysis/protocols/annexin-v-staining-flow-cytometry.html]
- 75. Apoptosis Protocols - USF Health - University of South Florida [https://health.usf.edu/medicine/corefacilities/flowcytometry/methods-apoptosis]
- 76. Annexin V PI Staining Guide for Apoptosis Detection [https://www.bosterbio.com/blog/post/methods/flow-cytometry-with-annexin-v-pi-staining?srsltid=AfmBOorOiYUfAu0TQ6cksCSbNY5mSICoQabFh_Zp-2Pd2i06hFaUSk4O]
- 77. Assays for Apoptosis and Autophagy—Section 15.5 [https://www.thermofisher.com/us/en/home/references/molecular-probes-the-handbook/assays-for-cell-viability-proliferation-and-function/assays-for-apoptosis.html]
- 78. Annexin V staining assay protocol for apoptosis [https://www.abcam.com/en-us/technical-resources/protocols/annexin-v-for-apoptosis]
- 79. Caspase-Independent Regulated Necrosis Pathways as ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7921719/]
- 80. Analyzing Cell Death by Nuclear Staining with Hoechst ... [https://pubmed.ncbi.nlm.nih.gov/27587774/]
- 81. How to Detect Key Features of Apoptosis? [https://www.enzo.com/note/how-to-detect-key-features-of-apoptosis/]
- 82. Programmed cell death detection methods - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC9308588/]
- 83. An update to DNA ladder assay for apoptosis detection [https://pmc.ncbi.nlm.nih.gov/articles/PMC4401164/]
- 84. A quantitative real-time approach for discriminating ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5253725/]
- 85. Guidelines for Regulated Cell Death Assays: A Systematic ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2021.634690/full]
- 86. Activation and Caspase-mediated Inhibition of PARP [https://pmc.ncbi.nlm.nih.gov/articles/PMC99613/]
- 87. Old, new and emerging functions of caspases | Cell Death ... [https://www.nature.com/articles/cdd2014216]
- 88. North America Apoptosis Assay Market Trends 2025–2034 [https://www.gminsights.com/industry-analysis/north-america-apoptosis-assay-market]
- 89. Visualization and detection of live and apoptotic cells with ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4654871/]
- 90. Apoptosis, Oxidative Stress, and Pathway Modulation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12566667/]
- 91. The Quantitative-Phase Dynamics of Apoptosis and Lytic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6994697/]
- 92. Restoring apoptosis in breast cancer via peptide mediated ... [https://www.nature.com/articles/s41598-025-24772-4]
- 93. Cell death in tumor microenvironment: an insight for exploiting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11894105/]
- 94. Imaging flow cytometry: a primer - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC10468826/]