Decoding Cell Death: A Comprehensive Guide to the Morphological Features of Apoptosis vs. Necrosis for Biomedical Research
Accurately distinguishing between apoptosis and necrosis is fundamental in biomedical research, influencing everything from basic mechanistic studies to the evaluation of anticancer therapies.
Decoding Cell Death: A Comprehensive Guide to the Morphological Features of Apoptosis vs. Necrosis for Biomedical Research
Abstract
Accurately distinguishing between apoptosis and necrosis is fundamental in biomedical research, influencing everything from basic mechanistic studies to the evaluation of anticancer therapies. This article provides a comprehensive resource for researchers and drug development professionals, detailing the specific morphological hallmarks of each process. It explores foundational concepts, advanced label-free imaging techniques like Full-Field Optical Coherence Tomography (FF-OCT), common challenges in interpretation, and validation strategies. By synthesizing classical knowledge with cutting-edge methodologies, this guide aims to enhance the precision of cell death analysis in experimental and clinical settings, ultimately supporting more reliable drug discovery and development.
The Core Blueprint: Defining the Classical Morphological Hallmarks of Apoptosis and Necrosis
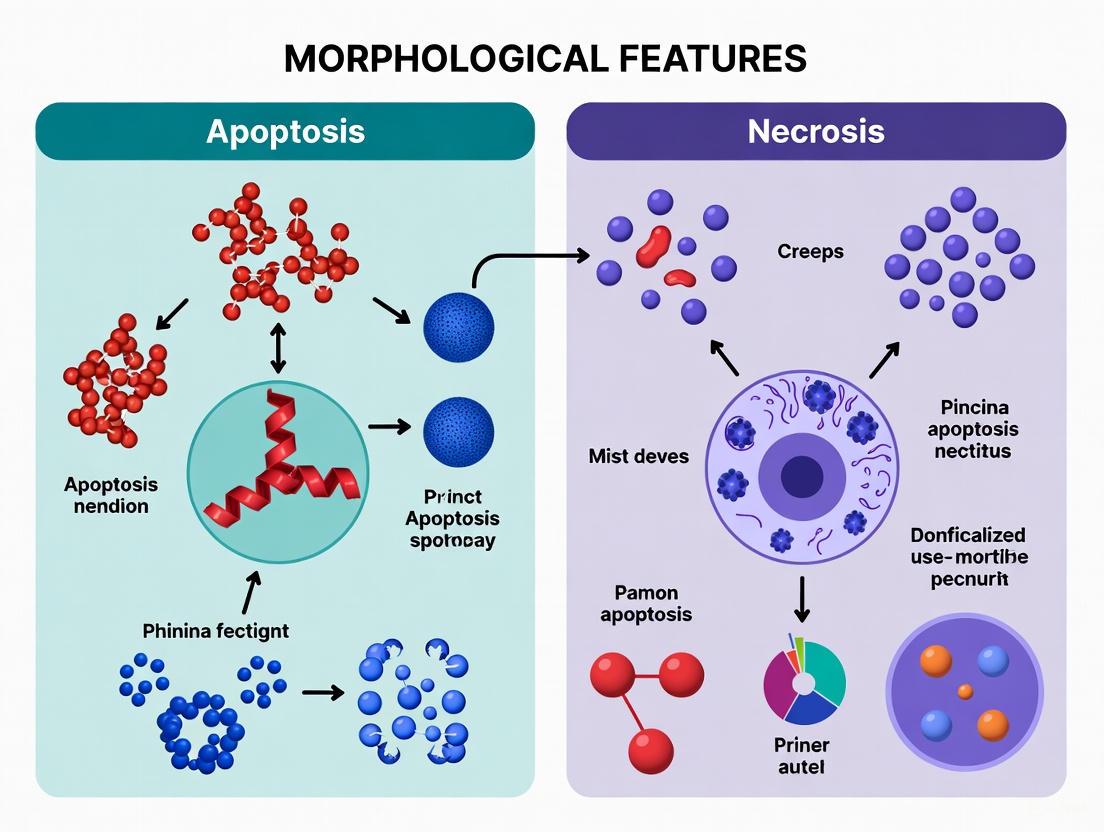
The precise identification of programmed cell death, or apoptosis, relies fundamentally on recognizing its unique morphological features, which stand in stark contrast to those of accidental cell death, or necrosis. These distinct morphological signatures are not merely descriptive; they are direct reflections of underlying biochemical pathways and have profound implications for tissue homeostasis and disease pathology. Apoptosis is a genetically regulated, energy-dependent process characterized by a tightly orchestrated series of structural changes, including cell shrinkage, membrane blebbing, and formation of apoptotic bodies, all occurring without triggering an inflammatory response [1] [2]. In contrast, necrosis is an unregulated, passive process resulting from overwhelming external injury, leading to cell swelling, membrane rupture, and spillage of intracellular contents that incites inflammation [1] [3]. This guide provides a detailed, evidence-based comparison of these morphological hallmarks, underpinned by experimental data and methodologies essential for researchers and drug development professionals.
Comparative Morphology: Apoptosis vs. Necrosis
The following table summarizes the key morphological and physiological differences that form the basis for distinguishing between apoptosis and necrosis in experimental settings.
Table 1: Comprehensive Comparison of Apoptotic and Necrotic Cell Death
| Feature | Apoptosis | Necrosis |
|---|---|---|
| Basic Nature | Programmed, regulated, energy-dependent [1] | Accidental, unregulated, energy-independent [3] |
| Cellular Trigger | Physiological or mild pathological signals; developmental cues [2] [4] | Extreme physical/chemical injury; toxins, trauma, infection [1] [5] |
| Cell Size and Shape | Cell shrinkage, cytoplasmic condensation [2] [6] | Cell swelling (oncosis) [2] |
| Plasma Membrane | Blebbing with intact integrity; formation of apoptotic bodies [1] [4] | Loss of integrity; rupture and leakage of cellular contents [1] [3] |
| Nuclear Changes | Chromatin condensation (pyknosis), nuclear fragmentation (karyorrhexis) [2] [6] | Karyolysis (nuclear dissolution); pyknosis and karyorrhexis can also occur [2] |
| Organelles | Generally intact, though mitochondria may leak contents [1] | Swelling and disintegration of ER, mitochondria, and lysosomes [1] [2] |
| DNA Fragmentation | Endonuclease-cleaved into orderly fragments (DNA laddering) [5] | Random, diffuse degradation [5] |
| Phagocytic Clearance | Rapid engulfment of apoptotic bodies by macrophages/neighboring cells [2] [4] | Cell lysis; phagocytosis of debris, often incomplete [5] |
| Tissue Response | Affects individual cells; no inflammatory response [1] [2] | Affects groups of contiguous cells; prominent inflammatory response [1] [2] |
| Key Biomarkers | Caspase activation, phosphatidylserine externalization [3] [5] | Loss of ion homeostasis, ATP depletion [1] |
Visualizing the Key Signaling Pathways
The morphological changes in apoptosis are directly executed by molecular pathways. The following diagram illustrates the two principal routes of apoptosis induction.
Experimental Imaging and Assessment Protocols
High-Resolution Imaging with FF-OCT
Advanced label-free imaging techniques like Full-Field Optical Coherence Tomography (FF-OCT) allow for real-time, high-resolution monitoring of apoptotic morphology.
- Principle: FF-OCT is an interferometric technique that uses a broadband light source to generate high-resolution, cross-sectional images of cells based on light scattering, without requiring stains or labels [7].
- Protocol:
- Cell Preparation: Culture HeLa cells (or other relevant cell line) as a monolayer under standard conditions (e.g., DMEM, 37°C, 5% CO₂) [7].
- Induction of Death:
- Image Acquisition: Use a custom-built time-domain FF-OCT system with a halogen light source (e.g., center wavelength 650 nm) and high-NA water immersion objectives (e.g., 40x, NA=0.8). Initiate imaging immediately post-treatment and acquire images continuously at set intervals (e.g., every 20 minutes for up to 3 hours) [7].
- Data Analysis: Reconstruct 3D surface topography from en face (x-y) cross-sectional data stacks. Analyze morphological parameters such as cell volume, membrane texture (blebbing), and adhesion structure [7].
- Key Observations:
- Apoptotic Cells: Show progressive cell contraction, formation of echinoid spines and membrane blebs, and filopodia reorganization. The cell membrane remains intact throughout [7].
- Necrotic Cells: Exhibit rapid membrane rupture, abrupt loss of adhesion structure, and leakage of intracellular contents, leading to a sudden loss of structural definition [7].
Classical Staining and Microscopy Techniques
Despite technological advances, traditional staining methods combined with microscopy remain a cornerstone for morphological assessment.
- Light Microscopy with H&E Staining:
- Protocol: Fix cells or tissue sections, stain with hematoxylin (nuclei blue) and eosin (cytoplasm pink). Examine under a light microscope [2] [6].
- Observation: Apoptotic cells appear as round/oval masses with dark eosinophilic (pink) cytoplasm and dense purple nuclear chromatin fragments. They often affect single cells or small clusters [2].
- Fluorescence Microscopy with Hoechst/DAPI:
- Protocol: Incubate live or fixed cells with membrane-permeable DNA-binding dyes like Hoechst 33342 (2-5 μg/mL) or DAPI. Examine under a fluorescence microscope with UV excitation [6].
- Observation: Viable nuclei show diffuse, dim staining. Apoptotic nuclei show bright, condensed, and fragmented chromatin, often aggregated at the nuclear periphery [6].
- Electron Microscopy:
- Protocol: Fix cells with glutaraldehyde and osmium tetroxide, dehydrate, embed in resin, section, and stain with heavy metals (e.g., uranyl acetate) [6].
- Observation: Provides the highest resolution view of subcellular changes, such as chromatin margination, intact organelles in apoptosis, and swollen organelles in necrosis [2] [6].
The following diagram illustrates a typical workflow for the morphological assessment of cell death.
Table 2: Key Research Reagents for Apoptosis and Necrosis Studies
| Reagent / Kit | Primary Function / Application | Key Experimental Notes |
|---|---|---|
| Doxorubicin [7] | Chemical inducer of intrinsic apoptosis via DNA intercalation and Topoisomerase II inhibition. | Used at ~5 μmol/L for in vitro apoptosis induction in HeLa cells. |
| Ethanol [7] | Chemical inducer of necrosis via protein denaturation and membrane disruption. | High concentrations (e.g., 99%) used for rapid, unregulated cell death. |
| Anti-Fas Antibody [8] | Activator of the extrinsic apoptosis pathway by clustering the Fas death receptor. | Used in Jurkat T-cell models to study receptor-mediated apoptosis. |
| Hoechst 33342 / DAPI [6] | Cell-permeable fluorescent DNA dyes for nuclear staining and visualization of chromatin condensation. | Excitation ~350 nm (UV), Emission ~461 nm (blue). Apoptotic nuclei show bright, condensed fluorescence. |
| Caspase-3 Antibody [5] | Immunodetection of cleaved/active caspase-3, a key executioner protease and gold-standard apoptotic biomarker. | Used in IHC, WB, and IF to confirm apoptosis commitment. |
| BAX Antibody [5] | Detects the pro-apoptotic protein BAX, which translocates to mitochondria during intrinsic apoptosis. | Indicator of mitochondrial pathway engagement. |
| Annexin V Assays | Binds to phosphatidylserine (PS) exposed on the outer leaflet of the apoptotic cell membrane. | Often used in flow cytometry with Propidium Iodide (PI) to distinguish early apoptotic (Annexin V+/PI-) from necrotic (Annexin V+/PI+) cells. |
| Dead Cell Removal Kits [1] | Magnetic or buoyancy-based separation of dead cells (often with compromised membranes) from live cell populations. | Improves purity and accuracy of downstream assays in cell isolation workflows. |
Critical Considerations and the Apoptosis-Necrosis Continuum
While the morphological distinctions between apoptosis and necrosis are foundational, the reality in experimental and pathological contexts is often more complex. Researchers must be aware that:
- The apoptosis-necrosis continuum: The same initial insult can cause apoptosis at low doses and necrosis at high doses [2]. Furthermore, features of both processes can coexist in the same tissue or even the same cell, a phenomenon sometimes called "apoptotic necrosis" or "necrapoptosis" [2] [6].
- Shared biochemical network: Apoptosis and necrosis are now understood to represent morphological expressions of a shared biochemical network. Key factors like the availability of caspases and intracellular ATP can determine the mode of death; for instance, if ATP is depleted during an apoptotic stimulus, the cell may undergo secondary necrosis [2].
- Emerging forms of programmed necrosis: Regulated forms of cell death that morphologically resemble necrosis, such as necroptosis, have been identified. Like apoptosis, it is genetically controlled, but it results in cell swelling and membrane rupture, similar to necrosis [1] [3] [5].
- Cell shrinkage is separable from apoptosis: Intriguing experimental evidence shows that under specific ionic conditions (sodium-substituted medium), cells can undergo all biochemical hallmarks of apoptosis, including caspase activation and DNA fragmentation, without shrinking, and instead swell [8]. This demonstrates that ion fluxes, not shrinkage itself, are critical for the death process.
Cell death is a fundamental biological process, and its accurate characterization is crucial for understanding disease pathogenesis and developing therapeutic strategies. Within this field, the morphological differentiation between apoptosis and necrosis provides critical insights into the mechanism and consequences of cell death. Necrosis is defined as an uncontrolled form of cell death triggered by external factors such as injury, trauma, infection, or toxins, leading to a cascade of distinctive morphological events [9] [10]. In contrast to programmed cell death pathways, necrosis is characterized by its unregulated nature and its tendency to trigger inflammatory responses [11] [10].
This guide provides a detailed, objective comparison of the morphological features of necrosis, with a specific focus on swelling, oncosis, and membrane rupture. It is structured within the broader thesis of differentiating apoptosis from necrosis, supplying researchers and drug development professionals with consolidated experimental data, validated protocols, and essential research tools for precise cell death identification.
Core Morphological Features of Necrosis
The progression of necrosis follows a characteristic sequence of events, beginning with cellular swelling and culminating in the complete loss of membrane integrity. The table below summarizes the key morphological stages and their functional consequences.
Table 1: Characteristic Morphological Events in Necrosis
| Morphological Event | Description | Functional Consequence |
|---|---|---|
| Cellular Swelling (Oncosis) | The cytoplasm and mitochondria swell up, leading to cell enlargement [9]. | Disruption of osmotic pressure balance and ion homeostasis [11]. |
| Plasma Membrane Rupture | The cell membrane loses integrity and ruptures (cell lysis) [9] [11]. | Leakage of intracellular contents into the extracellular space [9]. |
| Organelle Swelling | Organelles, including the ER and mitochondria, swell and disintegrate [9] [11]. | Cessation of organelle function and cellular metabolism [9]. |
| Inflammatory Response | Not a morphological feature of the cell per se, but a direct result of the release of cellular debris [11]. | Activation of immune cells, leading to inflammation and potential damage to surrounding tissues [9] [10]. |
The term "oncosis" is used specifically to describe the pre-lethal swelling process that precedes membrane rupture in necrosis [11]. This swelling is driven by the failure of ion pumps in the plasma membrane, leading to an influx of water and electrolytes. The loss of membrane integrity is a hallmark event, distinguishing necrosis from apoptosis, where membrane integrity is typically maintained until the final stages [9].
Objective Morphological Comparison: Necrosis vs. Apoptosis
Accurate differentiation between necrosis and apoptosis is a cornerstone of cell death research. The following table provides a side-by-side comparison of their defining characteristics, based on established experimental observations.
Table 2: Direct Comparison of Necrosis and Apoptosis Morphology
| Feature | Necrosis | Apoptosis |
|---|---|---|
| Cellular Process | Accidental, unregulated cell death from external injury [9]. | Programmed, controlled "cellular suicide" [9]. |
| Cell Morphology | Cytoplasm and mitochondria swell; cell undergoes lysis [9]. | Cell shrinks and condenses; cytoplasm dehydrates [9]. |
| Membrane Integrity | Lost; membrane ruptures and becomes highly permeable [9] [11]. | Largely maintained; membrane blebs but does not rupture [9]. |
| Organelle Behavior | Organelles swell and disintegrate [9]. | Organelles remain largely intact and functional until late stages [9]. |
| Fate of Cellular Contents | Released into extracellular space [9]. | Packaged into apoptotic bodies for phagocytosis [9]. |
| Inflammatory Response | Almost always triggered due to leaked contents [9] [11]. | Typically, no inflammation [9]. |
| Scope of Effect | Often affects contiguous groups of cells [9]. | Localized to individual cells [9]. |
The diagrams below visualize the distinct morphological pathways of necrosis and apoptosis, and a typical experimental workflow for their differentiation.
Cell Death Morphology: Necrosis Pathway
Workflow for Differentiating Cell Death
Experimental Data and Methodologies for Necrosis Detection
High-Resolution Imaging of Necrotic Morphology
Advanced label-free imaging techniques like Full-Field Optical Coherence Tomography (FF-OCT) allow for high-resolution, real-time observation of necrotic morphology.
Experimental Protocol [7]:
- Cell Preparation: HeLa cells are cultured as a monolayer in DMEM under standard conditions (37°C, 5% CO₂).
- Necrosis Induction: Cells are treated with 99% ethanol to induce nonspecific, rapid cellular damage leading to necrosis. Ethanol penetrates the lipid bilayer, disrupts membrane integrity, and denatures proteins.
- FF-OCT Imaging: A custom-built time-domain FF-OCT system with a broadband halogen light source is used. Imaging begins immediately post-treatment and continues at 20-minute intervals for up to 180 minutes.
- Data Analysis: 3D surface topography maps are reconstructed from the acquired tomographic image stacks to visualize and quantify morphological changes.
Key Observations [7]: FF-OCT effectively visualized the hallmark features of ethanol-induced necrosis, including rapid membrane rupture, intracellular content leakage, and an abrupt loss of adhesion structures.
Fluorescent Staining and Automated Quantification
Fluorescent staining kits provide a biochemical basis for differentiating cell death states and can be scaled for high-throughput analysis.
Experimental Protocol (Annexin V-CY3TM Kit) [12]:
- Cell Culture and Treatment: HeLa cells are seeded and allowed to adhere for 24 hours. Necrosis is induced, for example, via photodynamic treatment.
- Staining: Cells are simultaneously incubated with two probes: Annexin-Cy3.18 (AnnCy3), which binds to phosphatidylserine on the outer membrane, and 6-Carboxyfluorescein diacetate (6-CFDA), a viability dye hydrolyzed by intracellular esterases in live cells.
- Imaging: Cells are imaged using a confocal microscope with filters set for 6-CF (ex 495/em 520 nm) and AnnCy3 (ex 550/em 570 nm).
- Automated Analysis: The ApoNecV macro for the Fiji platform can automate the quantification. It performs background subtraction and deconvolution before classifying cells based on fluorescence:
- Viable cells: Green fluorescence (6-CF) only.
- Apoptotic cells: Both green (6-CF) and red (AnnCy3) fluorescence.
- Necrotic cells: Red fluorescence (AnnCy3) only, due to loss of membrane integrity and leakage of green 6-CF.
Key Observations [12]: This method reliably distinguishes necrotic cells (red-only signal) based on their compromised membrane, a defining feature absent in early apoptosis.
The Scientist's Toolkit: Essential Research Reagents
The following table lists key reagents and tools essential for conducting research on necrotic cell death.
Table 3: Key Reagents for Necrosis and Cell Death Research
| Research Reagent / Tool | Function and Application in Research |
|---|---|
| Annexin V-CY3TM Apoptosis Detection Kit | A standard kit using Annexin-Cy3 and 6-CFDA to distinguish viable, apoptotic, and necrotic cell populations via fluorescence microscopy or flow cytometry [12]. |
| Full-Field Optical Coherence Tomography (FF-OCT) | A label-free, non-invasive imaging technique for high-resolution 3D visualization of dynamic morphological changes like swelling and membrane rupture in single living cells [7]. |
| Propidium Iodide (PI) | A DNA stain that cannot cross intact membranes. It is used to label necrotic cells with compromised membrane integrity, often in conjunction with other markers [13]. |
| Lactate Dehydrogenase (LDH) Assay Kit | A colorimetric assay that measures LDH enzyme released from the cytosol upon plasma membrane rupture, providing a quantitative measure of necrotic cell death [13]. |
| Caspase Inhibitors (e.g., Z-VAD-FMK) | Pan-caspase inhibitors used experimentally to confirm a death process is caspase-independent, helping to rule out apoptosis and suggesting alternative pathways like necrosis [11]. |
| Chemical Inducers (e.g., Ethanol, H₂O₂) | Used to experimentally induce necrotic cell death in vitro. Ethanol (99%) causes rapid membrane damage and protein denaturation [7]. |
The morphological signature of necrosis—characterized by swelling (oncosis), organelle disintegration, and ultimate plasma membrane rupture—serves as a clear and reliable identifier for this inflammatory form of cell death. The experimental data and methodologies detailed in this guide, from high-resolution FF-OCT to accessible fluorescent staining and automated analysis, provide a robust framework for researchers to accurately distinguish necrosis from apoptosis. Mastery of these morphological distinctions and the associated toolkit is indispensable for advancing our understanding of disease mechanisms, assessing the efficacy of therapeutics, and ultimately developing safer and more effective drugs.
The microscopic examination of nuclear morphology remains a cornerstone in the fundamental classification of cell death pathways. Within the context of apoptosis and necrosis research, specific nuclear changes provide critical diagnostic markers that differentiate controlled, programmed cell death from uncontrolled, accidental death [2]. Chromatin condensation is an early and defining event in the process of programmed cell death, or apoptosis, representing an active, energy-dependent process [2]. This condensation progresses to pyknosis, characterized by nuclear shrinkage and hypercondensation of chromatin [14]. In contrast, karyolysis describes the complete dissolution of the nuclear structure and chromatin, typically following a necrotic pathway [15]. The precise identification and differentiation of these nuclear changes are not merely academic; they are essential for researchers and drug development professionals in accurately interpreting cellular responses to toxic insults, chemotherapeutic agents, and other disease-modifying therapies. This guide provides a structured comparison of these nuclear phenomena, supported by experimental data and methodologies relevant to modern cell biology research.
Comparative Morphology and Mechanisms
The journey of a dying cell is vividly reflected in the transformation of its nucleus. The distinct morphological pathways observed in apoptosis and necrosis offer a visual language for classifying cell death.
Apoptotic Nuclear Changes: A Controlled Demolition
Apoptosis, or programmed cell death, is a highly regulated process. Its nuclear changes are orderly and specific [2]:
- Chromatin Condensation and Pyknosis: The process begins with marginalization of chromatin, where it moves to the inner periphery of the nuclear membrane, forming a characteristic ring-like structure [15]. This is followed by pyknosis, the irreversible shrinkage and hypercondensation of the entire nucleus [14]. This stage is mediated by caspases (caspase-3 and -6), which cleave nuclear structural proteins like lamins and activate factors such as Acinus to initiate condensation [14] [15].
- Karyorrhexis: Following pyknosis, the condensed nucleus fragments into discrete particles, a process known as karyorrhexis [14] [2]. These nuclear fragments, along with intact organelles and cytoplasm, are then packaged into apoptotic bodies for efficient phagocytosis by neighboring cells, preventing an inflammatory response [2].
Necrotic Nuclear Changes: A Catastrophic Failure
Necrosis, often resulting from severe cellular injury, follows a more chaotic and disruptive course [16] [2]:
- Pyknosis in Necrosis: While also featuring pyknosis, the mechanism in necrosis differs and is termed anucleolytic pyknosis [14]. It involves cellular swelling, separation of the nuclear membrane from chromatin, and subsequent collapse of both structures. A key regulator is the phosphorylation of the barrier-to-autointegration factor (BAF), which disrupts the tethering of chromatin to the nuclear membrane [14].
- Karyolysis: This is the terminal stage of necrotic nuclear death, characterized by the complete dissolution of nuclear material [15]. The chromatin is degraded and digested, often due to the unspecific action of nucleases, leading to the "fading away" of the nucleus [15]. This occurs concurrently with a loss of plasma membrane integrity, causing the release of intracellular contents and provoking a strong inflammatory reaction [16] [2].
Table 1: Comparative Analysis of Nuclear Changes in Cell Death
| Feature | Apoptotic Pyknosis | Necrotic Pyknosis | Karyolysis |
|---|---|---|---|
| Primary Cell Death Type | Apoptosis (Programmed) [14] | Necrosis (Accidental) [14] | Necrosis (Accidental) [15] |
| Overall Process | Ordered, controlled, and energy-dependent [2] | Disordered, uncontrolled, and passive [2] | Passive dissolution [15] |
| Nuclear Morphology | Chromatin condensation → Nuclear fragmentation (Karyorrhexis) [2] | Nuclear shrinkage → Collapse of nuclear structure [14] | Complete dissolution of the nucleus [15] |
| Key Molecular Regulators | Caspase-3, Caspase-6, Acinus, CAD/DFF40 [14] [15] | Phosphorylated BAF, PLA2, AIF (in some models) [14] [15] | Lysosomal and exogenous nucleases (e.g., from Kupffer cells) [15] |
| Cellular Context | Cell shrinkage, membrane blebbing, formation of apoptotic bodies [16] [2] | Cell swelling (oncosis), organelle disruption, membrane rupture [16] [2] | Always follows necrotic pyknosis; occurs in a lytic cellular environment [15] |
| Inflammatory Response | None (anti-inflammatory) [2] | Significant (pro-inflammatory) [16] | Significant (pro-inflammatory) [16] |
Visualizing the Pathways of Nuclear Demise
The following diagram illustrates the sequence of key nuclear events in apoptosis and necrosis, highlighting the divergent paths from initial chromatin condensation to the final states of karyorrhexis or karyolysis.
Key Experimental Models and Data
Translating morphological observations into quantifiable data requires robust experimental models and assays. The following table summarizes key experimental findings that delineate apoptotic and necrotic cell death.
Table 2: Experimental Data Differentiating Apoptosis and Necrosis
| Experimental Model | Treatment / Inducer | Key Nuclear Morphology Observed | Primary Cell Death Type | Supporting Biochemical Evidence |
|---|---|---|---|---|
| Hepatic Ischemia-Reperfusion (Mouse) [17] | 45 min ischemia, 1-24h reperfusion | Predominant necrosis; TUNEL-positive cells showed karyorrhexis | Necrosis | High plasma ALT, miR-122, FK18, HMGB1; minimal caspase-3 activity and CK18 [17] |
| HepG2 Cell Line [18] | Anti-CD95 Antibody | Chromatin condensation, DNA fragmentation | Apoptosis | Cytochrome c release, caspase activation, specific protein cleavage [18] |
| HepG2 Cell Line [18] | Menadione | No DNA fragmentation | Necrosis | Cytochrome c release but no caspase activation; ATP depletion; inhibition reversed by catalase [18] |
| U251 Cell Line (Real-Time Imaging) [19] | Doxorubicin | Apoptotic morphology with subsequent shift to necrosis | Apoptosis → Secondary Necrosis | Caspase activation (FRET ratio change) followed by loss of FRET probe and mitochondrial fluorescence [19] |
| U251 Cell Line (Real-Time Imaging) [19] | Valinomycin / H₂O₂ | Necrotic morphology without caspase activation | Primary Necrosis | Loss of FRET probe without prior ratio change; retention of mitochondrial fluorescence [19] |
Detailed Experimental Protocol: Real-Time Discrimination
A sophisticated live-cell imaging method for distinguishing apoptosis and necrosis at a single-cell level is described by [19]. The protocol can be summarized as follows:
Cell Line Engineering: Stable expression of two genetically encoded probes in the target cell line (e.g., neuroblastoma U251 cells):
- A FRET-based caspase sensor (ECFP and EYFP linked by a DEVD caspase-cleavable sequence).
- A non-soluble fluorescent marker targeted to mitochondria (e.g., Mito-DsRed).
Treatment and Imaging: Cells are exposed to death-inducing stimuli (e.g., doxorubicin for apoptosis, H₂O₂ for necrosis) and subjected to real-time, time-lapse imaging using wide-field fluorescence or confocal microscopy.
Quantitative Discrimination and Data Analysis:
- Live Cells: Exhibit intact FRET probe (no ratio change) and retained mitochondrial red fluorescence.
- Apoptotic Cells: Show caspase activation, visualized by a loss of FRET (increase in ECFP/EYFP ratio), while retaining mitochondrial DsRed fluorescence.
- Necrotic Cells: Lose the soluble cytosolic FRET probe due to membrane permeabilization (no FRET ratio change) but continue to retain the mitochondrial DsRed fluorescence for a prolonged period.
This method allows for the sensitive and confirmatory real-time quantification of both primary necrosis and secondary necrosis (the lytic phase following apoptosis).
The Scientist's Toolkit: Essential Reagents for Cell Death Research
The following table catalogs key reagents and their applications in studying nuclear changes and cell death mechanisms.
Table 3: Research Reagent Solutions for Cell Death Studies
| Reagent / Assay | Function / Target | Application in Cell Death Research |
|---|---|---|
| Caspase-3 Antibody [20] | Detects activated/cleaved caspase-3 | Key biomarker for identifying apoptotic cells via IHC/IF; gold standard for apoptosis confirmation [3] [20]. |
| BAX Antibody [20] | Detects conformational change in BAX protein | Marker for intrinsic apoptosis pathway activation; indicates mitochondrial membrane pore formation [20]. |
| TUNEL Assay [14] [17] | Labels DNA strand breaks (3'-OH ends) | Detects DNA fragmentation. Note: Can be positive in both apoptosis and necrosis; requires morphological correlation [17]. |
| Annexin V / Propidium Iodide (PI) [19] | Binds phosphatidylserine (PS) / intercalates into DNA | Flow cytometry assay to distinguish early apoptotic (Annexin V+/PI-), late apoptotic/necrotic (Annexin V+/PI+), and necrotic (Annexin V-/PI+) cells. |
| Fixable Viability Dyes [16] | Covalently bind amine groups in dead cells | Flow cytometry-based dead cell removal; distinguishes cells based on membrane integrity. |
| Z-VD-fmk (Pan-Caspase Inhibitor) [17] | Irreversibly inhibits caspase activity | Used to confirm caspase-dependent apoptosis; can shift cell fate to necroptosis/necrosis [17] [18]. |
| AC-DEVD-AMC Fluorogenic Substrate [17] | Caspase-3/7 substrate | Measures caspase-3 activity in cell lysates or plasma; release of fluorescent AMC upon cleavage indicates apoptosis [17]. |
| Caspase Sensor Stable Cell Line [19] | FRET-based probe for caspase activation | Enables real-time, live-cell imaging and quantification of apoptosis as described in the experimental protocol [19]. |
Visualizing the Experimental Workflow
The following diagram outlines the logical workflow for designing an experiment to discriminate between apoptotic and necrotic cell death, integrating the reagents and methods from the toolkit.
Within the broader thesis of morphological feature specificity for apoptosis versus necrosis, the nuclear changes of pyknosis and karyolysis serve as fundamental, histologically accessible endpoints. While pyknosis can initiate both apoptotic and necrotic pathways, its context—preceding either organized karyorrhexis or catastrophic karyolysis—defines the ultimate nature of cell death. The experimental data and methodologies detailed herein provide a framework for researchers to move beyond simple observation to mechanistic insight. The choice of assay, from classic biomarker detection to advanced real-time imaging, depends on the specific research question, whether it involves screening compound toxicity, elucidating death signaling pathways, or characterizing novel cell death modalities. A precise understanding of these nuclear events remains indispensable for accurate interpretation of cellular fate in disease models and therapeutic interventions.
The integrity of the plasma membrane serves as a fundamental distinguishing feature between the two primary forms of cell death, apoptosis and necrosis. This characteristic not only defines their contrasting morphologies but also dictates their physiological consequences, particularly concerning inflammatory responses. The following table summarizes the core differences.
| Feature | Apoptosis | Necrosis (Accidental) |
|---|---|---|
| General Type | Regulated, Programmed Cell Death (RCD) [21] [22] | Accidental Cell Death (ACD) [21] [22] |
| Plasma Membrane Integrity | Maintained until late stages [23] [24] | Rapidly lost [23] [22] |
| Membrane Phenomena | Blebbing and formation of apoptotic bodies [23] [24] | Swelling and rupture (oncosis) [23] [22] |
| Key Process | Phosphatidylserine (PS) externalization as an "eat-me" signal [24] [3] | Non-selective release of intracellular contents [23] |
| Inflammatory Response | Typically non-inflammatory [23] [24] | Strongly pro-inflammatory [23] [24] |
| Primary Inducers | Physiological signals, developmental cues, mild damage [23] [22] | Extreme physical/chemical injury, toxins, ischemia [23] [25] |
Membrane Integrity: The Core Distinction
Apoptosis: A Sealed Demise
In apoptosis, the plasma membrane undergoes a carefully orchestrated transformation without immediately losing its role as a selective barrier. The cell exhibits membrane blebbing—the formation of outward bulges—driven by caspase-3-mediated activation of the ROCK1 kinase, which causes actomyosin contraction [24]. Crucially, the membrane's integrity is preserved during this blebbing [23]. A key biochemical event is the externalization of phosphatidylserine (PS), a phospholipid normally confined to the inner leaflet of the membrane. This surface exposure of PS acts as a critical "eat-me" signal for phagocytes, facilitating the prompt engulfment and disposal of the cell corpse without content leakage [24] [3]. Ultimately, the cell fragments into membrane-bound apoptotic bodies, which safely package cellular contents for phagocytosis [23] [24].
Necrosis: Loss of Barrier Function
Necrosis, in its classical accidental form, is characterized by an uncontrolled loss of plasma membrane integrity [22]. This failure of the barrier function is often preceded by cellular and organelle swelling (oncosis) [22]. The subsequent rupture leads to the non-selective release of intracellular components, such as lactate dehydrogenase (LDH), high mobility group box 1 (HMGB1), and other damage-associated molecular patterns (DAMPs) into the extracellular space [23] [24]. This unregulated spillage is a potent trigger for inflammation and immune cell recruitment, distinguishing it sharply from the typically non-inflammatory nature of apoptosis [23].
The diagram below illustrates the key morphological differences in the plasma membrane during each process.
Experimental Assessment of Membrane Integrity
Researchers employ specific reagents and assays to differentiate between apoptotic and necrotic cells based on membrane status. The cornerstone of this assessment is the differential permeability of fluorescent dyes.
Key Assays and Dye Permeability
A standard flow cytometry assay uses Annexin V in conjunction with membrane-impermeable DNA-binding dyes like Propidium Iodide (PI) or 7-AAD [26] [27].
- Annexin V+ / PI-: Early apoptotic cell (intact membrane, PS exposed).
- Annexin V+ / PI+: Late apoptotic or necrotic cell (compromised membrane).
- Annexin V- / PI+: Necrotic cell (membrane ruptured, no PS exposure).
Advanced research investigates specific pore-forming proteins. For example, during apoptosis-driven secondary necrosis, Gasdermin E (GSDME) can form pores in the plasma membrane. The influx and efflux of fluorescently-labeled dextrans of varying molecular weights can be monitored to characterize the functional size of these pores [28]. Dye choice is critical, as some, like YO-PRO-1 and TO-PRO-3, can enter early apoptotic cells via pannexin channels, complicating the interpretation of membrane integrity [26]. SYTOX dyes are often preferred for late-stage death as they are not associated with this early mechanism [26].
The experimental workflow for a basic membrane integrity assay is outlined below.
Quantitative Data from Experimental Models
The following table compiles key experimental findings on membrane permeability from selected studies.
| Experimental Model / Assay | Key Readout | Apoptosis Findings | Necrosis Findings | Citation |
|---|---|---|---|---|
| S49 Lymphoma Cells(Propidium Iodide Uptake) | Modest, gradual PI permeability | Yes, during early apoptosis | N/D | [27] |
| L929sAhFas Cells(Dextran Influx/Efflux) | Permeability to macromolecules | GSDME-dependent dextran influx during secondary necrosis | N/D | [28] |
| General Flow Cytometry(Annexin V/PI Staining) | Phosphatidylserine exposure & membrane integrity | Annexin V+ / PI- (early stage) | Annexin V- / PI+ | [26] [22] |
| General Morphology | Release of intracellular contents | Contents isolated in apoptotic bodies | Uncontrolled release of DAMPs | [23] [24] |
Underlying Signaling Pathways
The difference in membrane fate is determined by upstream molecular events.
Apoptosis: Caspase-Driven Regulation
Apoptosis proceeds via the extrinsic (death receptor) or intrinsic (mitochondrial) pathways, converging on the activation of caspase-3 [22] [3]. Caspase-3 cleaves specific substrates to orchestrate the disciplined dismantling of the cell:
- Cleavage of ROCK1: Triggers actomyosin contraction and membrane blebbing without immediate rupture [24].
- Externalization of Phosphatidylserine: Promotes immunologically silent phagocytosis [24].
- Cleavage of Gelsolin: Mediates disassembly of the actin cytoskeleton [24].
Necrosis and Programmed Necrosis: Direct Membrane Perforation
While accidental necrosis involves unregulated physical rupture, regulated forms like necroptosis and pyroptosis directly target the membrane.
- Necroptosis: Upon activation, the kinases RIPK1 and RIPK3 phosphorylate the pseudokinase MLKL. Phosphorylated MLKL oligomerizes and translocates to the plasma membrane, directly disrupting its integrity and causing lytic cell death [22].
- Pyroptosis: Inflammatory caspases (caspase-1/4/5) cleave Gasdermin D (GSDMD). Its N-terminal fragment forms large pores in the plasma membrane, leading to ion dysregulation, cell swelling, and release of pro-inflammatory cytokines [24] [29].
The Scientist's Toolkit: Key Research Reagents
The following reagents are essential for investigating plasma membrane integrity in cell death.
| Reagent / Assay | Primary Function | Key Considerations |
|---|---|---|
| Annexin V (conjugated to fluorophores) | Detects phosphatidylserine (PS) exposure on the outer leaflet of the plasma membrane, a hallmark of early apoptosis. | Requires calcium-containing buffer. Alone, it cannot distinguish between apoptotic and necrotic cells. |
| Propidium Iodide (PI) / 7-AAD | Membrane-impermeant DNA dyes that stain nuclei only in cells with compromised plasma membranes. | Used to identify late apoptotic and necrotic cells. Can be used in combination with Annexin V. |
| SYTOX Blue / Green | High-affinity, membrane-impermeant nucleic acid stains for identifying dead cells. | Provides a strong signal-to-noise ratio; not associated with early entry via pannexin channels [26]. |
| Antibodies against Cleaved Caspase-3 | Confirm the activation of the key executioner caspase in apoptosis. | Validates that cell death is occurring via the apoptotic pathway. |
| Antibodies against p-MLKL (Phospho-Mixed Lineage Kinase Domain-Like) | Detects the active form of MLKL, a key effector of necroptosis. | Specific marker for the regulated necroptotic pathway. |
| Lactate Dehydrogenase (LDH) Release Assay | Measures the activity of LDH enzyme released from the cytosol upon membrane rupture. | A standard colorimetric assay for quantifying cytotoxicity and lytic cell death. |
The integrity of the plasma membrane provides a clear and functionally decisive criterion for distinguishing apoptosis from necrosis. Apoptosis is a sealed, regulated process where membrane integrity is maintained to prevent inflammation, while necrosis is defined by a loss of membrane integrity that precipitates a potent inflammatory response. Advanced research continues to reveal the complex regulation behind these events, including the role of pore-forming proteins like Gasdermins and MLKL in programmed lytic cell death. A solid understanding of these principles and the associated experimental tools is indispensable for accurate interpretation of cell death phenomena in research and drug development.
Cell death is a fundamental physiological process, and its mode carries profound implications for organismal health. Among the various forms of cell death, apoptosis and necrosis represent two distinct mechanisms with opposing inflammatory outcomes. Apoptosis, known as programmed cell death, is a genetically regulated process that eliminates unwanted or damaged cells in a controlled manner without eliciting an inflammatory response, making it immunologically "silent" [30] [31]. In contrast, necrosis has traditionally been viewed as an unregulated, catastrophic form of cell death resulting from overwhelming external damage, characterized by cellular swelling and membrane rupture that leads to the release of intracellular contents and triggers a significant inflammatory reaction—rightfully earning its "alarming" designation [32] [33]. This fundamental difference in inflammatory potential stems from their unique morphological features, biochemical pathways, and physiological contexts, which we will explore through comparative morphological analysis, molecular mechanisms, and experimental approaches.
Morphological and Biochemical Hallmarks: A Comparative Analysis
The distinct inflammatory outcomes of apoptosis and necrosis are direct consequences of their structural disintegration processes, which can be observed through both microscopic and biochemical analyses.
Characteristic Features of Apoptosis
Apoptosis demonstrates a highly orchestrated series of morphological events that facilitate clean cell removal. The process initiates with cell shrinkage and loss of specialized cell-to-cell contacts [3] [33]. The nucleus undergoes characteristic chromatin condensation (pyknosis) followed by nuclear fragmentation (karyorrhexis) [30]. The cell membrane begins to bleb and form apoptotic bodies—small, membrane-bound vesicles containing intact organelles and nuclear fragments [3] [33]. Critically, the plasma membrane remains intact throughout this process, preventing the release of cellular contents [31]. Biochemically, apoptosis features phosphatidylserine externalization on the outer leaflet of the plasma membrane, which serves as an "eat me" signal for phagocytes [3] [33]. It also involves caspase activation and DNA cleavage into specific, regularly sized fragments (DNA laddering) [33] [30].
Characteristic Features of Necrosis
Necrosis presents a drastically different morphological pattern characterized by catastrophic cellular disintegration. The process begins with cell and organelle swelling (oncosis), including dilation of the endoplasmic reticulum and mitochondria [32] [33]. This is followed by plasma membrane rupture and subsequent release of intracellular contents—including organelles, proteins, and DNA fragments—into the extracellular space [32] [33]. The nucleus undergoes either dissolution (karyolysis) or irregular fragmentation without the organized pattern seen in apoptosis [32]. Unlike apoptosis, necrosis demonstrates random DNA degradation without specific fragment sizes, resulting in a "smear" pattern rather than a DNA ladder [33]. This loss of membrane integrity and release of cellular components directly triggers inflammatory responses [32] [33].
Table 1: Comparative Morphological and Biochemical Features of Apoptosis and Necrosis
| Feature | Apoptosis | Necrosis |
|---|---|---|
| Cell Size | Shrinkage | Swelling (oncosis) |
| Plasma Membrane | Intact with blebbing and apoptotic body formation; phosphatidylserine exposure | Rupture and disintegration |
| Organelles | Generally intact | Swelling and disruption |
| Nucleus | Chromatin condensation, pyknosis, karyorrhexis | Karyolysis, irregular fragmentation |
| DNA Fragmentation | Internucleosomal cleavage (DNA laddering) | Random degradation (DNA smear) |
| Inflammatory Response | None ("silent") | Significant ("alarming") |
| Energy Requirement | ATP-dependent | ATP-independent |
| Phagocytic Recognition | Efficient by neighboring cells | Inefficient; requires professional phagocytes |
Molecular Mechanisms: Signaling Pathway Dissection
The divergent inflammatory outcomes of apoptosis and necrosis are determined by their distinct molecular circuitry, which governs the cell death execution process.
The Apoptotic Signaling Cascade
Apoptosis proceeds through two main pathways that converge on caspase activation. The extrinsic pathway initiates through extracellular death ligands (e.g., FasL, TNF-α) binding to cell surface death receptors, leading to the formation of the Death-Inducing Signaling Complex (DISC) and activation of initiator caspase-8 [3] [30] [31]. The intrinsic pathway triggers in response to internal cellular damage (e.g., DNA damage, oxidative stress) through mitochondrial outer membrane permeabilization (MOMP), controlled by BCL-2 family proteins, resulting in cytochrome c release and formation of the apoptosome, which activates caspase-9 [3] [30] [31]. Both pathways converge to activate executioner caspases (caspase-3, -6, -7) that systematically cleave cellular substrates, leading to the characteristic morphological changes while maintaining membrane integrity [3] [30].
The Necrotic Signaling Cascade
While traditionally considered unregulated, certain forms of necrosis—specifically necroptosis—follow defined molecular pathways. Necroptosis can be initiated by death receptor activation (e.g., TNFR1) under conditions of caspase inhibition [3] [34]. This leads to the formation of a complex containing RIPK1 and RIPK3, known as the necrosome [34]. The necrosome phosphorylates the effector protein MLKL, causing it to oligomerize and translocate to the plasma membrane [33] [34]. MLKL oligomers form pores in the plasma membrane or activate ion channels, leading to membrane rupture, ion imbalance, and cellular swelling [34]. The subsequent release of damage-associated molecular patterns (DAMPs)—including HMGB1, ATP, and DNA fragments—activates immune cells and initiates robust inflammatory responses [32] [34].
Experimental Approaches: Discriminating Cell Death Modalities
Accurately distinguishing between apoptosis and necrosis is crucial for both research and clinical applications. Several well-established methodologies enable researchers to differentiate these processes based on their characteristic features.
Imaging-Based Discrimination Techniques
Advanced imaging technologies provide powerful tools for visualizing the distinct morphological changes associated with each cell death type. Full-field optical coherence tomography (FF-OCT) enables label-free, high-resolution visualization of apoptotic cells showing characteristic echinoid spine formation, cell contraction, membrane blebbing, and filopodia reorganization, while necrotic cells exhibit rapid membrane rupture, intracellular content leakage, and abrupt loss of adhesion structures [7]. Quantitative phase microscopy (QPM) allows non-invasive imaging of living cells by measuring phase shifts in transmitted light to map density distribution and refractive index variations within intracellular structures, revealing subtle structural differences between apoptotic and necrotic cells [7]. Live-cell imaging using FRET-based caspase sensors combined with organelle-targeted fluorescent proteins (e.g., Mito-DsRed) enables real-time discrimination at single-cell resolution—apoptotic cells show caspase activation (FRET loss) while retaining mitochondrial fluorescence, whereas necrotic cells lose the soluble FRET probe without caspase activation while maintaining organelle fluorescence [35].
Biochemical and Histological Assays
Multiple biochemical approaches capitalize on the distinct molecular events characterizing each cell death pathway. Annexin V/PI staining distinguishes early apoptotic cells (Annexin V+/PI-) with exposed phosphatidylserine but intact membranes from necrotic cells (Annexin V+/PI+) with compromised membrane integrity [17] [33]. Caspase activity assays using fluorogenic substrates (e.g., Ac-DEVD-AMC) or cleavage detection by Western blot specifically identify apoptotic cells [17] [30]. DNA fragmentation analysis through DNA laddering detection or TUNEL staining identifies apoptotic cells, though TUNEL can also stain necrotic cells with nonspecific DNA damage, requiring careful interpretation [17] [30]. Histological evaluation of tissue sections stained with hematoxylin and eosin (H&E) reveals characteristic patterns—apoptosis shows single-cell involvement with condensed nuclei and apoptotic bodies, while necrosis displays contiguous cells with swelling and loss of architecture [32] [17].
Table 2: Experimental Methods for Discriminating Apoptosis and Necrosis
| Method Category | Specific Assay | Apoptosis Detection | Necrosis Detection | Key Differentiating Feature |
|---|---|---|---|---|
| Imaging | FF-OCT | Cell shrinkage, membrane blebbing | Membrane rupture, content leakage | Membrane integrity during death process |
| Live-Cell Imaging | FRET-based caspase sensors + Mito-DsRed | Caspase activation with organelle retention | Loss of soluble probes with organelle retention | Caspase activation vs. membrane permeability |
| Flow Cytometry | Annexin V/PI staining | Annexin V+/PI- (early) | Annexin V+/PI+ | Membrane integrity with phosphatidylserine exposure |
| Biochemical | Caspase activity assays | Caspase-3/7, -8, or -9 activation | No caspase activation | Presence of specific protease activity |
| Molecular | DNA fragmentation analysis | DNA laddering pattern | Random DNA smear | Pattern of DNA degradation |
| Histological | H&E staining | Single cells, condensed nuclei, apoptotic bodies | Cell groups, swelling, architecture loss | Tissue pattern and nuclear morphology |
The Scientist's Toolkit: Essential Research Reagents and Methods
This section details crucial laboratory reagents and methodologies employed in the key studies cited throughout this article, providing researchers with practical experimental guidance.
Table 3: Essential Research Reagents and Methods for Cell Death Studies
| Reagent/Method | Application | Experimental Function | Key Considerations |
|---|---|---|---|
| Doxorubicin | Apoptosis induction | Chemotherapeutic agent that intercalates into DNA, causing double-strand breaks and activating p53 pathway [7] [35] | Typically used at 5 μmol/L concentration; effective in rapidly proliferating cells |
| Ethanol | Necrosis induction | Lipid solvent that disrupts membrane integrity and denatures proteins at high concentrations [7] | 99% concentration induces rapid necrotic death; effects are concentration-dependent |
| Z-VD-fmk | Caspase inhibition | Pan-caspase inhibitor used to distinguish caspase-dependent and independent death pathways [17] | Can shift cell fate from apoptosis to necroptosis when death receptors are engaged |
| Recombinant TNF-α | Death receptor activation | Cytokine that activates TNFR1, inducing either apoptosis or necroptosis depending on cellular context [34] | Outcome depends on downstream signaling modifiers (caspase activity, NF-κB activation) |
| Annexin V-FITC/PI | Flow cytometry detection | Dual staining distinguishes early apoptotic (FITC+/PI-) from necrotic (FITC+/PI+) cells [33] | Requires careful timing as late apoptotic cells become PI+ (secondary necrosis) |
| TUNEL Assay | DNA fragmentation detection | Labels DNA strand breaks in situ; identifies apoptotic and necrotic cells [17] [36] | Not specific for apoptosis; necrotic cells with random DNA damage also stain positive |
| Caspase-3 Antibody | Immunodetection | Detects caspase-3 cleavage/activation as specific apoptosis marker [33] | Cleaved caspase-3 is definitive apoptosis indicator; multiple commercial antibodies available |
| FRET-based Caspase Sensor | Live-cell imaging | Genetically encoded probe (ECFP-DEVD-EYFP) shows caspase activation by FRET loss [35] | Enables real-time, single-cell analysis of caspase activation dynamics |
The fundamental distinction between the "silent" nature of apoptosis and the "alarming" character of necrosis has profound implications for both physiological homeostasis and pathological conditions. Apoptosis serves as the primary mechanism for programmed cell elimination during development, tissue remodeling, and immune system regulation without provoking inflammation or tissue damage [30] [31]. In contrast, necrosis typically occurs under pathological conditions—such as ischemia, trauma, or infection—where it not only causes local tissue damage but also amplifies inflammatory responses through DAMP release [32] [34]. Understanding these differential inflammatory outcomes provides critical insights for therapeutic development, particularly in oncology where promoting apoptosis in cancer cells represents a key treatment strategy, while minimizing necrosis-associated inflammation can improve therapeutic outcomes [31] [35]. Future research continues to explore the complex interplay between these cell death pathways and their modulation for therapeutic benefit across diverse disease contexts.
Morphological and Functional Distinction: Necroptosis vs. Apoptosis
The classical view of cell death delineated a clear boundary: apoptosis was a programmed, controlled process, while necrosis was considered an unregulated, accidental death resulting from extreme injury or stress [37] [38]. The discovery of necroptosis fundamentally challenged this dichotomy, revealing a form of cell death that is genetically programmed yet exhibits a necrotic morphology [39] [40].
The table below provides a detailed comparison of the core characteristics of apoptosis and necroptosis, highlighting the distinct profile of necroptosis as a regulated pathway with inflammatory consequences.
Table 1: Comparative Analysis of Apoptosis and Necroptosis
| Feature | Apoptosis | Necroptosis |
|---|---|---|
| Regulation | Programmed, highly regulated [38] | Programmed and regulated [39] [40] |
| Morphology | Cell shrinkage, chromatin condensation, membrane blebbing, formation of apoptotic bodies [38] [41] | Cell and organelle swelling, plasma membrane rupture [42] [39] [40] |
| Membrane Integrity | Maintained until late stages (blebbing) [38] [41] | Lost, leading to permeabilization and rupture [42] [39] |
| Inflammatory Response | Typically none (non-immunogenic) [38] [41] | Strong, pro-inflammatory response [39] [40] [43] |
| Key Mediators | Caspases (e.g., Caspase-3, -8) [38] [41] [44] | RIPK1, RIPK3, MLKL [37] [42] [39] |
| Energy Dependence | ATP-dependent [41] | ATP-independent [41] |
| Primary Physiological Role | Developmental programming, tissue homeostasis [38] [41] | Host defense against pathogens, alternative death pathway when apoptosis is blocked [39] [41] |
Molecular Mechanisms and Key Signaling Pathways
Necroptosis is initiated by specific stimuli, most notably Tumor Necrosis Factor (TNF-α) binding to its receptor TNFR1, but also by Toll-like receptors (TLR3, TLR4), and viral sensors [39] [40]. The core regulatory machinery involves a cascade of protein interactions and phosphorylation events.
The following diagram illustrates the critical molecular switch at the heart of the TNF-α-induced necroptotic pathway.
Diagram Title: Molecular Switch Between Apoptosis and Necroptosis
Pathway Breakdown:
- Initiation: TNF-α binding to TNFR1 triggers the formation of a membrane-associated complex (Complex I) containing TRADD, TRAF2/5, and cellular Inhibitor of Apoptosis Proteins (cIAP1/2). Within this complex, RIPK1 is polyubiquitinated, leading to the activation of pro-survival NF-κB signaling [39] [40].
- The Switch: Deubiquitination of RIPK1 (e.g., by the enzyme CYLD) prompts the formation of a cytoplasmic complex. This complex includes FADD and procaspase-8. Active caspase-8 cleaves and inactivates RIPK1 and RIPK3, promoting apoptosis. However, when caspase-8 activity is inhibited—a common strategy employed by viruses or cancer cells—the cell defaults to necroptosis [39] [40].
- Necrosome Formation: In the absence of caspase-8 activity, RIPK1 and RIPK3 interact through their RIP Homotypic Interaction Motif (RHIM) domains, forming a structure known as the necrosome. This leads to auto- and trans-phosphorylation of RIPK3 [42] [39].
- Execution: Phosphorylated RIPK3 then recruits and phosphorylates the key executioner protein, Mixed Lineage Kinase Domain-Like (MLKL). This causes MLKL to oligomerize and translocate to the plasma membrane, where it integrates and forms pores, leading to membrane rupture, release of cellular contents, and the characteristic inflammatory response [42] [39] [40].
Experimental Approaches for Studying Necroptosis
Dissecting the necroptotic pathway relies on a combination of genetic, pharmacological, and biochemical techniques. The table below outlines key reagents and methodologies used to probe this form of cell death experimentally.
Table 2: Research Reagent Solutions for Necroptosis Investigation
| Research Tool | Type | Mechanism of Action / Function | Key Experimental Use |
|---|---|---|---|
| Necrostatin-1 (Nec-1) [37] | Small-molecule inhibitor | Potent and specific inhibitor of RIPK1 kinase activity. | Used to confirm RIPK1-dependent necroptosis in vitro and in vivo. A cornerstone tool for establishing the regulated nature of the process [37]. |
| GSK'872 / GSK'843 [37] | Small-molecule inhibitor | Selective inhibitor of RIPK3 kinase activity. | Used to inhibit downstream signaling from RIPK3, distinguishing RIPK1-dependent from RIPK1-independent necroptosis pathways [37]. |
| Necrosulfonamide [37] | Small-molecule inhibitor | Directly targets and inhibits human MLKL. | Blocks the final execution step of necroptosis, preventing plasma membrane rupture. Used to confirm MLKL's role as the effector protein [37]. |
| z-VAD-FMK [39] | Pan-caspase inhibitor | Irreversibly inhibits caspase activity. | Used experimentally to block apoptosis and create conditions permissive for necroptosis induction (e.g., in combination with TNF-α) [39]. |
| siRNA/shRNA | Genetic tool | Knocks down expression of specific target genes (e.g., RIPK1, RIPK3, MLKL). | Validates the essential role of specific proteins in the necroptotic pathway in a genetic loss-of-function context [42]. |
| Phospho-Specific Antibodies (e.g., anti-pMLKL) | Immunological reagent | Detects the phosphorylated, active form of MLKL. | Used in Western blotting and immunohistochemistry as a definitive biochemical marker for ongoing necroptosis [42]. |
Detailed Experimental Protocol: Inducing and Confirming Necroptosis in Vitro
A standard protocol for inducing and validating necroptosis in cell culture (e.g., in mouse fibroblast L929 or HT-29 cells) involves the following steps [37] [39]:
Stimulation: Treat cells with a combination of:
- TNF-α (e.g., 20-50 ng/mL): The primary death ligand.
- z-VAD-FMK (e.g., 20 µM): A pan-caspase inhibitor to block apoptosis.
- Smac mimetic (e.g., 100 nM): To degrade cIAP1/2 and promote complex II formation. This combination is often referred to as TSZ treatment.
Inhibition Control: Include control groups where the TSZ treatment is supplemented with specific inhibitors:
- Necrostatin-1 (10-30 µM) to inhibit RIPK1.
- GSK'872 (1 µM) to inhibit RIPK3. Cell viability in these groups, measured 16-24 hours post-treatment, should be significantly higher than in the TSZ-only group.
Cell Death Assessment:
- Viability Assays: Quantify cell death using methods like propidium iodide (PI) uptake followed by flow cytometry, or MTT/XTT assays. Necroptotic cells, with their compromised membranes, will be PI-positive.
- Morphological Analysis: Observe cells by phase-contrast microscopy for hallmarks of necrosis: swelling, plasma membrane blebbing (without fragmentation into apoptotic bodies), and eventual lysis.
Biochemical Confirmation:
- Western Blotting: Analyze cell lysates for key signaling events.
- Detect phosphorylation of RIPK3 and MLKL.
- Monitor cleavage of caspases (e.g., Caspase-3, -8) to confirm apoptosis inhibition by z-VAD.
- Western Blotting: Analyze cell lysates for key signaling events.
The following flowchart summarizes this multi-faceted experimental approach.
Diagram Title: Workflow for Necroptosis Induction and Validation
Necroptosis in Disease and Therapy: A Double-Edged Sword
The potent immunogenicity of necroptosis underpins its dual role in disease, functioning as both a protective mechanism and a pathogenic driver.
- Anti-Tumor Potential: Necroptosis induction is a promising strategy for overcoming apoptosis resistance in cancers. For instance, the prognostic signature of necroptosis-related genes like
CAMK2A,CHMP4C, andPYGBin Lung Squamous Cell Carcinoma (LUSC) highlights its clinical relevance [42]. Furthermore, nanomaterials are being engineered to specifically trigger necroptosis in tumor cells, offering a novel approach to cancer therapy [40]. - Pathological Inflammation: Conversely, aberrant necroptosis is implicated in the pathology of numerous inflammatory and degenerative diseases. The uncontrolled release of DAMPs can drive damaging inflammation in conditions such as ischemia-reperfusion injury (e.g., in heart attack and stroke), inflammatory bowel disease, and neurodegenerative disorders like Alzheimer's and Huntington's disease [37] [39] [43]. In these contexts, pharmacological inhibition of key nodes in the pathway (e.g., with Necrostatin-1) has shown protective effects in preclinical models [37].
The Scientist's Toolkit: Key Reagents for Necroptosis Research
The following table consolidates essential tools that form the backbone of experimental necroptosis research.
Table 3: Essential Research Reagents for Necroptosis Investigation
| Category | Reagent Examples | Primary Function |
|---|---|---|
| Inducers | TNF-α, TSZ combination, LPS + z-VAD, Poly(I:C) [37] [39] | Activate death receptors or TLRs to initiate the necroptotic signaling cascade. |
| RIPK1 Inhibitors | Necrostatin-1, Nec-1s, PN10 [37] | Specifically block RIPK1 kinase activity, used to confirm upstream involvement of RIPK1. |
| RIPK3 Inhibitors | GSK'840, GSK'843, GSK'872 [37] | Specifically block RIPK3 kinase activity, used to inhibit the core necrosome complex. |
| MLKL Inhibitors | Necrosulfonamide (human-specific) [37] | Blocks the pore-forming activity of MLKL, preventing the final step of membrane disruption. |
| Caspase Inhibitors | z-VAD-FMK, Q-VD-OPh [39] | Pan-caspase inhibitors used to create permissive conditions for necroptosis by blocking apoptosis. |
| Detection Tools | Anti-pMLKL antibodies, Propidium Iodide (PI), Viability dyes [42] | Used to biochemically confirm pathway activation (pMLKL) and assess loss of membrane integrity (PI). |
Beyond the Microscope: Advanced Techniques for Visualizing Cell Death Morphology
In biomedical research, particularly in drug development and toxicology, the ability to distinguish between apoptosis and necrosis is crucial. These two modes of cell death have distinct morphological features and physiological implications. Apoptosis is a genetically regulated, programmed process essential for development and tissue homeostasis, while necrosis represents an uncontrolled, pathological response to severe injury [45] [7]. Traditional methods for distinguishing these pathways often rely on fluorescent staining, which can alter native cell biology and preclude long-term live-cell imaging. Full-field optical coherence tomography (FF-OCT) has emerged as a powerful label-free imaging technique that enables non-invasive, high-resolution visualization of dynamic cellular processes without requiring contrast agents or sample fixation [45] [46] [7].
FF-OCT represents a significant advancement over conventional OCT by illuminating and detecting the entire field of view simultaneously using a two-dimensional camera, enabling rapid, en face imaging with isotropic resolution approaching 1 μm [46]. This technical approach provides several advantages for live-cell imaging: minimal photodamage, continuous monitoring of the same sample over extended periods, and the ability to reconstruct three-dimensional cellular architectures from optical sections [47]. When applied to the study of cell death mechanisms, FF-OCT captures the distinctive, dynamic morphological changes that characterize apoptotic and necrotic pathways, providing researchers with a powerful tool for drug toxicity testing, anticancer therapy evaluation, and regenerative medicine applications [45] [7].
Technical Foundations of FF-OCT for Live-Cell Imaging
Basic Principles and System Configurations
FF-OCT operates on the principles of low-coherence interferometry, typically implemented in a Linnik microscope configuration with identical high-numerical-aperture objectives in both reference and sample arms [45] [48]. A spatially incoherent light source, such as a halogen lamp or light-emitting diode (LED), provides illumination, with the broad spectral bandwidth determining the axial resolution of the system [48] [7]. When the optical path lengths of the reference and sample arms are matched within the coherence length of the source, interference occurs between light back-reflected from the reference mirror and back-scattered from intracellular structures within the sample [49] [48].
The resulting interferometric signals are detected simultaneously across the entire field of view using a high-speed megapixel camera [48]. In traditional FF-OCT systems, depth sectioning is achieved by phase-shifting the reference mirror using a piezoelectric transducer and processing multiple interferograms to reconstruct en face optical sections [50] [7]. More recent innovations include continuous-scanning FF-OCT, which eliminates the need for piezoelectric modulation by continuously translating the sample axially while acquiring images, then applying Fourier analysis to the temporal signal at each pixel to extract depth-resolved information [50]. This approach simplifies system design and enables faster volumetric imaging—typically acquiring a 400 μm volume within 100 seconds [50].
Dynamic FF-OCT and Metabolic Contrast
A significant advancement in FF-OCT technology is the development of dynamic FF-OCT (D-FFOCT), which analyzes temporal fluctuations in the interferometric signal to generate contrast based on intracellular motility and metabolic activity [46] [47] [48]. In D-FFOCT, a time series of interferograms (typically 500 images at 100 Hz) is recorded at each imaging position, capturing signal variations induced by natural dynamics within biological samples [48]. The frequency characteristics of these temporal variations are then computed through power spectral density analysis and color-coded, with transitions from red to blue indicating increasing signal variation speed, and brightness corresponding to variation magnitude [48].
This dynamic contrast mechanism is particularly valuable for distinguishing different cellular states and activities. Metabolic processes such as mitochondrial dynamics, vesicular transport, and membrane fluctuations generate distinctive signal patterns that can be quantified using algorithms including standard deviation (STD), logarithmic intensity variation (LIV), and OCT correlation decay speed (OCDS) analysis [46]. The integration of both structural and functional imaging capabilities within a single platform makes dual-mode FF-OCT systems particularly powerful for comprehensive cell state assessment, especially in the context of differentiating apoptosis from necrosis [48].
Experimental Methodology for Cell Death Imaging
Cell Preparation and Death Induction Protocols
To investigate apoptotic and necrotic pathways using FF-OCT, researchers typically employ established cell lines such as HeLa (human cervical cancer cells) cultured under standard conditions [45] [7]. For apoptosis induction, doxorubicin—an anthracycline chemotherapeutic agent that intercalates into DNA and inhibits topoisomerase II—is added to the culture medium at a final concentration of 5 μmol/L [45] [7]. This treatment triggers intracellular injury responses, including activation of the p53 pathway and increased reactive oxygen species production, leading to programmed cell death. For necrosis induction, cells are treated with 99% ethanol, which causes nonspecific cellular damage through membrane disruption, protein denaturation, and loss of ion homeostasis, resulting in uncontrolled cell death [45] [7]. FF-OCT imaging typically begins immediately after drug administration and continues at regular intervals (e.g., every 20 minutes) for up to 180 minutes to capture the dynamic morphological changes associated with each death pathway [7].
FF-OCT Imaging Setup and Parameters
Imaging is performed using a custom-built time-domain FF-OCT system with the following typical specifications [45] [7]:
- Light Source: Broadband halogen lamp (center wavelength: 650 nm, spectral width: 200 nm)
- Interferometer: Linnik-configured Michelson interferometer with identical 40× water-immersion objectives (NA: 0.8)
- Detection: CCD camera (1024 × 1024 pixels, 12-bit, 20 fps)
- Phase Shifting: Piezoelectric actuator attached to reference mirror (20 nm resolution)
- Axial Resolution: <1 μm (achieved through broad spectral bandwidth)
- Lateral Resolution: <1 μm (determined by objective NA)
For dynamic FF-OCT imaging, systems typically acquire 500 interferograms at a rate of 100 Hz at each depth position, with the entire volumetric acquisition process optimized through parallelized acquisition, data transfer, and processing to achieve a total time of approximately 5.12 seconds per D-FFOCT image [47].
Image Processing and Analysis Techniques
The processing of FF-OCT data involves several specialized algorithms to extract meaningful structural and dynamic information [50] [46] [48]:
- Structural FF-OCT: Uses phase-shifting interferometry (typically 4-phase modulation) to reconstruct en face optical sections by arithmetic combination of phase-shifted interferograms, effectively isolating the sample reflection information while suppressing background signals [7].
- Dynamic FF-OCT: Applies Fourier analysis to time-series data at each pixel to compute power spectral density, with subsequent color-coding of frequency characteristics. Additional processing steps may include singular value decomposition and adaptive threshold filtering to eliminate dynamic artifacts [48].
- 3D Reconstruction: Combines multiple en face sections into z-stacks using precise motorized staging, enabling three-dimensional visualization of cellular structures and surface topography mapping through identification of the depth of maximum intensity at each pixel position [7].
Table 1: Key Research Reagent Solutions for FF-OCT Cell Death Studies
| Reagent/Component | Function/Application | Specifications |
|---|---|---|
| HeLa Cell Line | Model system for apoptosis/necrosis studies | Human cervical cancer cells (KCLB-10002) |
| Doxorubicin | Apoptosis induction agent | 5 μmol/L in culture medium |
| Ethanol | Necrosis induction agent | 99% concentration |
| Water-Immersion Objectives | High-resolution imaging | 40×, NA=0.8, WD=3.3 mm |
| Broadband Light Source | Interferometry illumination | Halogen lamp, 650 nm center wavelength |
| Piezoelectric Actuator | Reference mirror modulation | 20 nm resolution |
Morphological Differentiation of Apoptosis and Necrosis
Characteristic Features of Apoptotic Cells
FF-OCT imaging reveals a distinct sequence of morphological changes during apoptosis, characterized by well-orchestrated structural modifications that maintain membrane integrity until late stages [45] [7]. The process typically begins with cell contraction and echinoid spine formation, where the cell undergoes shrinkage and develops spike-like protrusions from the surface [45] [7]. This is followed by membrane blebbing, characterized by the formation of bulging, blister-like structures on the cell surface as the cytoskeleton reorganizes and the plasma membrane detaches from underlying structures [45] [51] [7]. Concurrently, filopodia reorganization occurs, with fine cytoplasmic projections retracting or extending in a dynamic manner [7]. As apoptosis progresses, the cell fragments into apoptotic bodies—membrane-bound vesicles containing condensed cytoplasm and organelles—which are eventually phagocytosed by neighboring cells without triggering inflammation [52] [7]. Throughout this process, the nuclear envelope remains intact, and the cell maintains its ability to exclude vital dyes until the final stages [52].
Characteristic Features of Necrotic Cells
In contrast to the organized process of apoptosis, necrosis presents as a disorderly series of structural deteriorations resulting from catastrophic cellular damage [45] [7]. The most prominent feature is rapid membrane rupture, where the plasma membrane becomes compromised, leading to uncontrolled release of intracellular contents [45] [7]. This is accompanied by intracellular content leakage, as cytoplasmic components spill into the extracellular space, often triggering inflammatory responses in surrounding tissues [45] [7]. Necrotic cells typically exhibit cell swelling rather than contraction, as ion homeostasis collapses and osmotic balance is disrupted [52] [7]. There is also an abrupt loss of adhesion structures, causing detached cells to float freely in the culture medium [45] [7]. Unlike apoptosis, where organelles remain largely intact until late stages, necrosis involves widespread organelle degradation, including mitochondrial swelling and endoplasmic reticulum dilation, visible as loss of internal structural detail in FF-OCT images [7].
Table 2: Comparative Morphological Features of Apoptosis and Necrosis Visualized by FF-OCT
| Morphological Feature | Apoptosis | Necrosis |
|---|---|---|
| Cell Size | Contraction/SHRINKAGE | Swelling/INCREASE |
| Plasma Membrane | Intact until late stages; Membrane blebbing | Early rupture; Loss of integrity |
| Nuclear Morphology | Chromatin condensation; Nuclear fragmentation | Nonspecific degradation |
| Cytoplasmic Organelles | Initially intact; Sequestered in apoptotic bodies | Swelling; Generalized disruption |
| Cellular Adhesion | Gradual loss maintained until late stages | Abrupt and early loss |
| Inflammatory Response | None (phagocytosis by adjacent cells) | Marked (release of intracellular contents) |
| Filopodia | Reorganization | Disintegration |
| Time Course | Progressive (hours) | Rapid (minutes to hours) |
Experimental Data and Comparative Analysis
Quantitative Assessment of Morphological Changes
FF-OCT enables not only qualitative visualization but also quantitative analysis of morphological parameters during cell death. Using FF-OCT-based three-dimensional topographic mapping, researchers can track changes in cell volume, surface area, and height distribution with submicrometer resolution [7]. Apoptotic cells typically show a progressive decrease in volume (up to 30-50% reduction) accompanied by an initial increase in surface roughness due to membrane blebbing, followed by smoothing as the cell fragments into apoptotic bodies [7]. In contrast, necrotic cells demonstrate rapid volume increase (up to 50-100% expansion) followed by sudden collapse as the membrane ruptures [7]. The integration of interference reflection microscopy (IRM)-like imaging with FF-OCT provides additional quantitative data on cell-substrate adhesion, with apoptotic cells showing gradual detachment while necrotic cells exhibit abrupt loss of adhesion contacts [45] [7].
Temporal Dynamics of Cell Death Processes
The dynamic imaging capabilities of FF-OCT reveal significant differences in the timing and progression of apoptotic versus necrotic pathways. Apoptosis typically follows a slower, more synchronized time course, with initial morphological changes appearing 60-120 minutes after doxorubicin exposure and progressing over several hours [7]. Membrane blebbing generally peaks around 2-3 hours post-induction, followed by fragmentation into apoptotic bodies [7]. In contrast, ethanol-induced necrosis manifests within minutes, with membrane rupture and content leakage occurring within 30-60 minutes of treatment [7]. The real-time monitoring capability of FF-OCT allows researchers to precisely document these temporal patterns and identify transitional stages that might be missed with fixed-timepoint sampling methods.
Comparative Advantages of FF-OCT in Cell Death Research
Performance Relative to Alternative Imaging Modalities
When compared to other imaging techniques commonly used in cell death research, FF-OCT offers several distinct advantages that make it particularly suitable for distinguishing apoptosis from necrosis:
Versus Fluorescence Microscopy: FF-OCT provides label-free imaging that eliminates phototoxicity and photobleaching concerns, enabling longer-term observation of dynamic processes without altering native cell physiology [45] [47]. While fluorescence methods offer molecular specificity through targeted probes, they cannot reliably distinguish apoptosis from necrosis without multiple staining protocols that may themselves affect cell viability [7].
Versus Electron Microscopy: Although electron microscopy provides superior resolution, it requires sample fixation and sectioning, precluding real-time observation of dynamic processes [45] [7]. FF-OCT enables live-cell monitoring of the entire death process without introducing artifacts from chemical fixation or processing.
Versus Quantitative Phase Microscopy (QPM): While both are label-free techniques, FF-OCT provides better optical sectioning capabilities and more direct visualization of intracellular structures [7]. QPM images phase shifts in transmitted light and may struggle with low refractive index contrast, whereas FF-OCT detects back-scattered light with greater sensitivity to fine structural details [7].
Versus Conventional OCT: FF-OCT offers significantly higher resolution (approximately 1 μm versus 10 μm in conventional OCT) due to its use of high-NA objectives and full-field detection, enabling visualization of subcellular features essential for distinguishing apoptotic and necrotic morphologies [46] [48].
Applications in Drug Development and Toxicity Testing
The label-free, non-invasive nature of FF-OCT makes it particularly valuable for pharmaceutical applications, where it can monitor cell responses to candidate compounds in real-time without introducing artifacts from sample preparation [45] [7]. In drug toxicity testing, the ability to distinguish apoptosis from necrosis is critical, as these different death modes may indicate distinct mechanisms of toxicity and have different implications for tissue damage and inflammatory responses [45] [7]. FF-OCT can provide early indicators of compound-induced cytotoxicity before complete cell death occurs, through detection of subtle morphological changes and alterations in intracellular dynamics [46]. For anticancer therapy evaluation, the technique can determine whether chemotherapeutic agents are successfully inducing apoptosis in target cells while monitoring for off-target necrotic effects that could contribute to undesirable side effects [45] [7].
Full-field optical coherence tomography represents a significant advancement in label-free live-cell imaging, providing researchers with a powerful tool for distinguishing between apoptotic and necrotic cell death pathways based on their distinctive morphological signatures. The technique's ability to capture high-resolution, three-dimensional structural information while simultaneously monitoring dynamic intracellular processes enables comprehensive characterization of cell states without the need for contrast agents or sample fixation. As FF-OCT technology continues to evolve, with improvements in imaging speed, resolution, and analytical algorithms, its applications in drug development, toxicology assessment, and basic cell biology research are expected to expand significantly. The quantitative, non-invasive nature of FF-OCT imaging makes it particularly valuable for long-term studies of cellular responses to therapeutic agents, providing insights that bridge the gap between in vitro assays and in vivo outcomes.
Three-Dimensional Topographic Mapping of Cell Death Structures
In the fields of cell biology, toxicology, and drug development, accurately distinguishing between different modes of cell death is crucial for understanding compound mechanisms, assessing therapeutic efficacy, and evaluating toxicity. For decades, this discrimination has relied heavily on biochemical assays and fluorescent staining techniques. However, a paradigm shift is underway toward label-free, non-invasive imaging methods that can visualize and quantify the distinct three-dimensional morphological changes associated with apoptosis and necrosis without potentially artifactual interference from stains or labels.
This guide provides a comparative analysis of advanced imaging platforms enabling three-dimensional topographic mapping of cell death structures, with a focus on the morphological features specific to apoptosis versus necrosis. We objectively evaluate the performance of leading technologies, present supporting experimental data, and detail the methodologies required to implement these approaches in the research laboratory.
Comparative Analysis of 3D Imaging Platforms
The following table compares the key technologies currently employed for the 3D morphological characterization of cell death.
Table 1: Comparison of 3D Imaging Platforms for Cell Death Analysis
| Technology | Spatial Resolution | Key Morphological Features for Apoptosis | Key Morphological Features for Necrosis | Label-Free | Live-Cell Capable | Throughput |
|---|---|---|---|---|---|---|
| Full-Field Optical Coherence Tomography (FF-OCT) [7] [45] | <1 μm (axial and transverse) | Cell contraction, membrane blebbing, filopodia reorganization, echinoid spine formation [7] [45] | Rapid membrane rupture, intracellular content leakage, abrupt loss of adhesion structure [7] [45] | Yes [7] [45] | Yes [7] [45] | Medium |
| Holographic Tomography (HT) [53] | Sub-cellular (based on RI) | Cell fragmentation, nucleus condensation into circular shape [53] | Cell swelling [53] | Yes [53] | Yes (with specific systems) | Medium |
| Digital Holographic Microscopy (DHM) with Deep Learning [54] | N/A (Phase shift) | Cell shrinkage, membrane blebbing [54] | Cell rounding and swelling [54] | Yes [54] | Yes [54] | High (>10,000 cells) [54] |
| Confocal Microscopy [55] | ~200-300 nm (lateral) | Nuclear fragmentation and condensation, membrane blebbing [55] | Not specifically detailed | No (requires staining) [55] | Possible with vital dyes | Low |
Experimental Protocols for Key Assays
FF-OCT for Single-Cell Death Visualization
This protocol is adapted from studies using FF-OCT to monitor doxorubicin-induced apoptosis and ethanol-induced necrosis in HeLa cells [7] [45].
- Cell Preparation: Culture HeLa cells as a monolayer in DMEM under standard conditions (37°C, 5% CO₂).
- Cell Death Induction:
- Apoptosis: Treat cells with 5 μmol/L doxorubicin in 1.5 mL culture medium. Doxorubicin intercalates into DNA, causing double-strand breaks and activating p53-mediated apoptosis [7] [45].
- Necrosis: Treat cells with 99% ethanol. Ethanol disrupts the phospholipid bilayer, denatures proteins, and causes rapid membrane rupture [7] [45].
- FF-OCT Imaging:
- System: Use a custom-built time-domain FF-OCT system with a Linnik-configured Michelson interferometer.
- Light Source: Broadband halogen lamp (center wavelength: 650 nm, spectral width: 200 nm).
- Objectives: Identical 40× water-immersion objectives (NA: 0.8) in reference and sample arms.
- Image Acquisition: Initiate imaging immediately post-treatment, acquiring en face (x-y) cross-sectional images at 20-minute intervals for up to 180 minutes. Use a CCD camera for detection.
- 3D Reconstruction: Stack z-axis tomographic images using a motorized stage. Reconstruct 3D surface topography by mapping the depth of maximum reflected intensity for each pixel [7] [45].
Holographic Tomography for 3D Refractive Index Mapping
This protocol is adapted from a study investigating autophagy, apoptosis, and ferroptosis in SH-SY5Y neuroblastoma cells [53].
- Cell Culture and Death Induction:
- Autophagy: Transfect cells with GFP-LC3 plasmid and culture in serum-free medium for 16 hours.
- Apoptosis: Expose cells to UV light for 1 hour, followed by a 30-minute incubation.
- Ferroptosis: Treat cells with 10 mM sodium iodate for 24 hours.
- Validation Assays:
- Perform Annexin V/PI staining and flow cytometry for apoptotic cells.
- Use PI staining for ferroptotic cells.
- Perform immunofluorescence imaging with LysoTracker Red and DAPI for autophagic cells.
- HT Imaging and Analysis:
- Acquire 3D refractive index (RI) distributions of fixed cells using a holographic tomography system.
- Calculate quantitative morphological parameters (projected area, height, volume, nucleo-cytoplasmic ratio) from the 3D RI model [53].
Digital Holographic Microscopy with Deep Learning
This protocol is adapted from a study discriminating apoptosis and necroptosis in L929sAhFas cells [54].
- Cell Death Induction:
- Apoptosis: Treat cells with an anti-Fas antibody to trigger the extrinsic apoptosis pathway.
- Necroptosis: Treat cells with mTNF to induce necroptosis.
- DHM Image Acquisition:
- Acquire quantitative phase images of cells using a transmission DHM setup without fluorescent labels.
- Deep Learning Analysis:
- Data Preparation: Crop single cells from the full-field-of-view DHM images.
- Anomaly Detection: Apply a supervised anomaly detection (SAD) filter to remove alive cells from the apoptosis and necroptosis datasets.
- Model Training and Classification: Use a transfer learning approach with a pre-trained VGG-19 convolutional neural network. Fine-tune the model to classify cells as alive, apoptotic, or necroptotic based on their phase images [54].
Signaling Pathways and Experimental Workflows
The diagram below illustrates the core signaling pathways of apoptosis and necrosis, highlighting key morphological features that can be captured via 3D imaging.
Diagram 1: Signaling Pathways in Cell Death. This diagram outlines the major pathways leading to apoptotic and necrotic cell death, culminating in their distinct morphological features.
The following diagram illustrates a generalized workflow for conducting 3D topographic mapping of cell death structures, from sample preparation to quantitative analysis.
Diagram 2: Experimental Workflow for 3D Cell Death Mapping. This diagram outlines the key steps from initial cell preparation to final data interpretation in a 3D morphological analysis pipeline.
The Scientist's Toolkit: Essential Research Reagents and Materials
The following table details key reagents, cell lines, and materials essential for conducting experiments in 3D topographic mapping of cell death.
Table 2: Essential Research Reagents and Materials for Cell Death Morphology Studies
| Item | Specification / Example | Function / Application | Reference |
|---|---|---|---|
| Model Cell Line | HeLa (Human cervical cancer) | Standard model for studying apoptosis and necrosis [7] [45] | [7] [45] |
| Model Cell Line | SH-SY5Y (Human neuroblastoma) | Model for studying apoptosis, autophagy, and ferroptosis [53] | [53] |
| Model Cell Line | L929sAhFas (Murine fibrosarcoma) | Model for discriminating apoptosis and necroptosis in the same cellular context [54] | [54] |
| Apoptosis Inducer | Doxorubicin (5 μmol/L) | Chemotherapeutic agent; induces intrinsic apoptosis by intercalating into DNA and inhibiting topoisomerase II [7] [45] | [7] [45] |
| Necrosis Inducer | Ethanol (99%) | Induces accidental necrosis by disrupting the phospholipid bilayer and denaturing proteins [7] [45] | [7] [45] |
| Necroptosis Inducer | mTNF (Murine Tumor Necrosis Factor) | Activates the RIPK1/RIPK3/MLKL pathway to induce regulated necrosis [54] | [54] |
| Fluorescent Apoptosis Marker | Annexin V (with FITC or other conjugates) | Binds to phosphatidylserine exposed on the outer leaflet of the apoptotic cell membrane [55] | [55] |
| Nucleic Acid Stain | Syto-61, Hoechst 33342, DAPI | Stains nuclear material; allows visualization of chromatin condensation and nuclear fragmentation [53] [55] | [53] [55] |
| Viability/Death Stain | Propidium Iodide (PI) | Membrane-impermeant dye that stains DNA in cells with compromised plasma membranes (necrotic/late apoptotic) [53] | [53] |
| Caspase Activity Probe | FRET-based DEVD probe (e.g., CFP-DEVD-YFP) | Genetically encoded biosensor for detecting caspase activation in live cells [35] | [35] |
| Inhibitor (Apoptosis) | zVAD-fmk (pan-caspase inhibitor) | Blocks caspase activity, thereby inhibiting apoptosis [54] | [54] |
| Inhibitor (Necroptosis) | Necrostatin-1s (Nec-1s) | Selective inhibitor of RIPK1, thereby blocking necroptosis [54] | [54] |
Discussion and Concluding Remarks
The advent of high-resolution, label-free 3D imaging technologies has fundamentally enhanced our ability to discriminate between different modes of cell death based on definitive morphological criteria. While apoptosis is consistently characterized by cell shrinkage, membrane blebbing, and nuclear condensation, necrosis is identified by cell swelling, plasma membrane rupture, and organelle disintegration [7] [45] [3].
The choice of imaging platform involves strategic trade-offs. FF-OCT excels in providing high-resolution, label-free, real-time tomography of single cells [7] [45]. In contrast, Holographic Tomography provides quantitative 3D refractive index distributions, offering insights into subcellular mass density changes [53]. When combined with deep learning, as in DHM, the analysis achieves high-throughput, automated classification capabilities that can process tens of thousands of cells with remarkable accuracy [54].
For researchers, the critical takeaway is that these 3D morphological techniques are not merely replacements for traditional biochemical assays but are complementary tools that provide a direct, unbiased window into the physical manifestations of cell death. Integrating these morphological analyses with established biochemical markers creates a more robust and conclusive framework for cell death discrimination, ultimately accelerating research in drug discovery, toxicology, and fundamental cell biology.
Interference Reflection Microscopy (IRM) for Assessing Cell-Substrate Detachment
The precise distinction between different forms of cell death, particularly apoptosis and necrosis, is fundamental to biomedical research in areas ranging from cancer treatment to toxicology. These cell death pathways exhibit distinct morphological characteristics that serve as crucial diagnostic markers [3]. Apoptosis, a genetically regulated and energy-dependent process, is characterized by cell shrinkage, chromatin condensation, nuclear fragmentation, and plasma membrane blebbing, ultimately leading to the formation of apoptotic bodies that are phagocytosed without triggering inflammation [3] [56]. In contrast, necrosis represents an accidental and catastrophic form of cell death characterized by cellular swelling, plasma membrane rupture, and the release of intracellular contents that provoke a significant inflammatory response [3] [45].
A critical yet often overlooked morphological event in cell death is the alteration in cell-substrate adhesion. During apoptosis, cells undergo a controlled process of detachment from their underlying substrate, while necrotic cells typically maintain adhesion until membrane integrity is completely lost [45]. These differential adhesion dynamics provide a valuable morphological parameter for distinguishing between these two cell death pathways. Interference Reflection Microscopy (IRM) has emerged as a powerful, non-invasive technique capable of quantitatively monitoring these nanoscale changes in cell-substrate separation during cell death processes, offering researchers a unique window into the initial stages of cellular detachment [45] [57].
Fundamental Principles of IRM
Interference Reflection Microscopy is a specialized light microscopy technique that enables the visualization of cell-substrate contacts with nanometer-scale axial resolution. The fundamental principle underlying IRM is interferometry, where light waves reflected from two closely spaced surfaces interfere with each other [57]. In a typical IRM configuration for cell biology applications, these two surfaces are the culture substrate (e.g., glass coverslip) and the ventral membrane of the adherent cell [57].
When monochromatic light is directed through the substrate toward the cell, portions of the light beam are reflected from both the substrate-liquid interface and the liquid-cell membrane interface. These reflected beams recombine and interfere, producing an interference pattern that is highly sensitive to the separation distance between the cell membrane and substrate [57]. The resulting intensity (I) at each point in the image follows the relationship:
I ≈ I₁ + I₂ + 2√(I₁I₂)cos(4πnd/λ + π)
Where I₁ and I₂ represent the intensities of the two reflected beams, n is the refractive index of the medium, d is the cell-substrate separation distance, and λ is the wavelength of the illuminating light [57]. The additional π phase shift accounts for the phase change occurring at the higher refractive index surface. This relationship enables IRM to detect distance changes as small as 5-10 nm, making it exceptionally sensitive to early adhesion changes [57] [58].
IRM System Configuration
A typical IRM setup is built around an inverted microscope equipped with specific optical components optimized for interference contrast imaging [57]. Essential components include:
- A monochromatic light source (laser or filtered lamp) with high spatial coherence
- A high-numerical aperture objective (typically oil-immersion, NA ≥1.3) to maximize light collection
- Specialized filter cubes containing beam splitters and emission filters optimized for the illumination wavelength
- A high-sensitivity camera (CMOS or CCD) for capturing low-light interference patterns
For dynamic studies of cell death processes, the system is often integrated with an environmental chamber to maintain physiological conditions (37°C, 5% CO₂) throughout time-lapse imaging sessions [57]. The exceptional sensitivity of IRM necessitates rigorous vibration isolation to prevent artifacts from mechanical disturbances that could obscure the subtle nanometer-scale motions of the cell membrane [57].
Comparative Analysis of Cell Death Morphology
Distinct Adhesion Dynamics in Apoptosis vs. Necrosis
The application of IRM to study cell death has revealed fundamentally different patterns of cell-substrate detachment in apoptosis versus necrosis, providing critical discriminatory morphological criteria.
Apoptotic Detachment Characteristics
In apoptosis, IRM reveals a gradual, progressive increase in cell-substrate separation distance that correlates with other hallmark morphological events [45]. This detachment begins early in the apoptotic process, often preceding overt membrane blebbing and nuclear fragmentation. The IRM signature shows a steady diminution of close contacts (appearing as dark areas in IRM images) accompanied by a corresponding expansion of regions with greater separation distances (appearing as brighter areas) [45]. This pattern reflects the controlled disassembly of focal adhesions and reorganization of the actin cytoskeleton that occurs during programmed cell death. The detachment process typically unfolds over tens of minutes to hours, mirroring the gradual progression of apoptosis [45].
Necrotic Adhesion Profile
In contrast, necrosis exhibits a dramatically different IRM profile characterized by maintained adhesion until the final stages of cell death [45]. IRM imaging shows that close contacts remain largely intact until the moment of plasma membrane rupture. Rather than the controlled retraction seen in apoptosis, necrotic cells display abrupt loss of adhesion structures coinciding with catastrophic membrane failure [45]. This is followed by rapid collapse of the normal interference pattern as intracellular contents leak into the extracellular space, creating a homogeneous bright field due to the loss of optical interfaces [45]. The entire process occurs rapidly, often within minutes of the initial insult, reflecting the uncontrolled nature of necrotic death.
Comprehensive Morphological Comparison
The table below summarizes the key morphological differences between apoptosis and necrosis, highlighting parameters detectable by various imaging methods:
Table 1: Comparative Morphological Features of Apoptosis and Necrosis
| Parameter | Apoptosis | Necrosis |
|---|---|---|
| Cell Volume | Decreased (cell shrinkage) [3] | Increased (cell swelling) [3] |
| Plasma Membrane | Blebbing, intact, phospholipid scrambling [3] [56] | Rupture, loss of integrity [3] |
| Nuclear Changes | Chromatin condensation, fragmentation [3] [56] | Karyolysis, nuclear swelling [3] |
| Cell-Substrate Adhesion (IRM) | Gradual, progressive detachment [45] | Maintained until membrane rupture [45] |
| Mitochondria | Condensed, cytochrome c release [3] | Swelling, membrane disruption [3] |
| Inflammatory Response | None (phagocytic clearance) [3] | Significant inflammation [3] |
| Kinetics | Gradual (hours) [3] | Rapid (minutes) [3] |
Experimental Protocols for IRM in Cell Death Studies
Sample Preparation and Imaging
Proper sample preparation is essential for successful IRM analysis of cell death processes. The following protocol has been optimized for studying apoptosis and necrosis in adherent cell cultures:
Substrate Preparation: Plate cells onto high-quality glass-bottom dishes (e.g., 35mm μ-Dishes) that have been thoroughly cleaned to minimize autofluorescence and imaging artifacts. For optimal adhesion studies, substrates may be coated with appropriate extracellular matrix proteins (e.g., fibronectin, collagen) at physiologically relevant densities [57].
Cell Seeding: Seed cells at subconfluent density (typically 30-50% confluence) to allow clear visualization of individual cells and their adhesion patterns. Allow cells to adhere and spread for 12-24 hours under standard culture conditions before experimentation [57].
Cell Death Induction:
IRM Imaging:
- Replace culture medium with phenol-red free medium supplemented with 20mM HEPES buffer to maintain pH during imaging without CO₂ control [57].
- Mount the dish on the microscope stage equipped with a temperature controller set to 37°C.
- Using a 100× oil-immersion objective (NA ≥1.3), locate cells of interest and acquire baseline IRM images.
- Collect time-lapse images at 30-second to 2-minute intervals for up to 3-6 hours to capture the dynamics of cell detachment [45] [57].
Image Processing and Data Analysis
Raw IRM images require processing to extract quantitative information about cell-substrate separation:
Background Subtraction: Acquire a reference image of a cell-free region and subtract this background from all experimental images to correct for uneven illumination [57].
Region of Interest (ROI) Selection: Define ROIs corresponding to different adhesion states (close contacts, intermediate distances, etc.) based on intensity thresholds [57].
Quantitative Analysis:
- Measure the total area of close contacts (darkest regions in IRM, corresponding to separations <30 nm) over time.
- Calculate the average separation distance across the entire cell-substrate interface.
- Track the redistribution of adhesion patterns during cell death progression.
Data Correlation: Correlate IRM adhesion metrics with other cell death markers (e.g., membrane blebbing, Annexin V binding) acquired through parallel imaging modalities [59].
Diagram 1: IRM Experimental Workflow for Cell Death Studies
Comparison with Alternative Cell Death Assessment Methods
Technical Comparison of Detection Methods
IRM represents one of several methodologies available for detecting and characterizing cell death. The table below provides a comprehensive comparison of IRM with other commonly used techniques:
Table 2: Comparison of Cell Death Detection Methodologies
| Method | Principle | Detection Parameters | Advantages | Limitations |
|---|---|---|---|---|
| IRM | Light interference at cell-substrate interface [57] | Nanoscale adhesion changes, early detachment [45] [57] | Label-free, non-invasive, high axial resolution, live-cell compatible [45] [57] | Limited to adherent cells, requires specialized optics, lower throughput [57] |
| Annexin V Staining | Phosphatidylserine externalization [56] | Loss of membrane asymmetry (early apoptosis) [56] | Early apoptosis detection, can distinguish apoptosis/necrosis with viability dyes [56] | Requires staining, membrane permeabilization in late apoptosis/necrosis [56] |
| DNA Fragmentation (TUNEL) | DNA strand break labeling [56] | Late-stage apoptosis (DNA fragmentation) [56] | High specificity for apoptosis, works on fixed tissue [56] | Late-stage detection only, cannot detect early apoptosis [56] |
| Caspase Activity Assays | Protease activity measurement [56] | Caspase activation (mid-apoptosis) [56] | Specific for apoptosis, various formats available [56] | Caspase activation doesn't always commit to apoptosis [56] |
| Electron Microscopy | Ultrastructural visualization [56] | Subcellular morphology, organelle changes [3] [56] | High resolution, definitive morphological assessment [56] | Fixed samples only, labor-intensive, low throughput [56] |
| Full-Field OCT | Interferometric tomography [45] [7] | 3D cellular morphology, membrane dynamics [45] [7] | Label-free, 3D structural information, high resolution [45] [7] | Complex instrumentation, computationally intensive [45] |
Advantages of IRM in Specific Research Contexts
IRM provides unique advantages for particular experimental scenarios:
Early Detection Capability: IRM can detect adhesion changes that often precede other biochemical markers of apoptosis, providing earlier detection than Annexin V binding or caspase activation assays [45] [56].
Non-invasive Monitoring: As a label-free technique, IRM enables long-term observation of the same cells throughout death progression without introducing potential artifacts from fluorescent labels or inhibitors [45] [57].
High Temporal Resolution: The ability to capture images at seconds-to-minutes intervals makes IRM ideal for tracking the rapid dynamics of cell detachment, unlike endpoint assays like TUNEL or electron microscopy [57] [59].
Correlative Studies: IRM can be readily combined with other imaging modalities (e.g., fluorescence microscopy) to correlate adhesion dynamics with molecular events in the same cells [57].
Essential Research Reagent Solutions
The table below outlines key reagents and materials essential for implementing IRM studies of cell death:
Table 3: Essential Research Reagents for IRM Cell Death Studies
| Reagent/Material | Function/Application | Examples/Specifications |
|---|---|---|
| Glass-bottom Culture Dishes | High-resolution imaging substrate | 35mm μ-Dishes, #1.5 coverglass thickness (0.16-0.19mm) [57] |
| Extracellular Matrix Proteins | Promote cell adhesion for imaging | Fibronectin, collagen, poly-L-lysine coatings [57] |
| Apoptosis Inducers | Experimental induction of apoptosis | Doxorubicin (5 μmol/L), Etoposide (10 μmol/L) [45] [59] |
| Necrosis Inducers | Experimental induction of necrosis | Ethanol (70-99%), hydrogen peroxide [45] |
| Phenol-red Free Medium | Reduce background autofluorescence during imaging | DMEM or RPMI without phenol red [59] |
| HEPES Buffer | Maintain pH during extended imaging without CO₂ control | 20mM concentration in imaging medium [57] |
| Immersion Oil | Optimize light collection for high-resolution IRM | Type B/F, matched to objective specifications [57] |
| Viability Stains | Confirm cell death mode when combined with IRM | Propidium iodide, Annexin V conjugates [56] |
Interference Reflection Microscopy represents a powerful methodology for assessing cell-substrate detachment during cell death processes, providing unique insights into the adhesion dynamics that differentiate apoptosis from necrosis. Its ability to detect nanometer-scale separation changes in living cells without chemical labels makes it particularly valuable for kinetic studies of death progression. When integrated with complementary techniques that probe biochemical events, IRM contributes to a comprehensive understanding of cell death mechanisms. The continued refinement of IRM methodologies, particularly its combination with advanced optical techniques like full-field OCT [45] [7], promises to further enhance our capability to resolve the subtle morphological alterations that characterize different cell death pathways, ultimately advancing drug development and toxicological assessment.
Nuclear Magnetic Resonance (NMR) Spectroscopy for Biochemical Fingerprinting of Apoptosis
Within the broader context of morphological feature specificity for apoptosis versus necrosis research, distinguishing these distinct modes of cell death is crucial for assessing anticancer drug efficacy and understanding disease pathology. Apoptosis, a genetically regulated and programmed process, features characteristic biochemical events that occur long before morphological changes become visible under conventional microscopy. In contrast, necrosis represents an uncontrolled and catastrophic cell death triggered by physicochemical injury, leading to membrane rupture and inflammatory responses [7]. The ability to accurately differentiate between these processes at an early stage significantly impacts biomedical fields, including pathological diagnostics, drug response assessment, and cancer treatment development [60] [7].
Nuclear Magnetic Resonance (NMR) spectroscopy has emerged as a powerful, non-destructive analytical technique capable of detecting subtle metabolic alterations associated with apoptosis through biochemical fingerprinting. Unlike morphological techniques that visualize structural changes, NMR identifies specific metabolic signatures and intensity ratios that serve as reliable biomarkers for programmed cell death. This approach provides researchers with a quantitative method to distinguish apoptosis from necrosis early in the death process, enabling more accurate assessment of treatment efficacy in drug development pipelines [60] [61].
NMR-Based Metabolic Signatures of Apoptosis Versus Necrosis
Key Metabolic Biomarkers in Apoptosis
NMR spectroscopy detects specific metabolic changes in apoptotic cells that serve as reliable fingerprints for this programmed cell death pathway. Research across multiple cell lines, including human cervical carcinoma (HeLa) cells and normal human lung (MRC-5) cells, has consistently identified characteristic metabolic alterations during apoptosis.
A pivotal finding across studies is the significant increase in methylene (CH₂) resonance intensity relative to methyl (CH₃) resonance intensity during apoptosis. This CH₂/CH₃ ratio demonstrates a strong positive correlation with the percentage of apoptotic cells in a sample (r² = 0.965, P < 0.003 in HeLa cells; r² = 0.9868, P < 0.01 in MRC-5 cells) [60] [61]. This increase in methylene resonance becomes detectable as early as 6 hours after apoptosis induction, preceding most morphological changes [60]. Additionally, the choline resonance (at 3.2 ppm) remains stable during apoptosis but is abolished in necrosis, providing a clear distinguishing feature between these cell death pathways [61].
Table 1: Key Metabolic Biomarkers for Apoptosis Detection via ¹H-NMR Spectroscopy
| Metabolic Parameter | Spectral Change in Apoptosis | Time of Onset | Correlation with Apoptosis | Biological Significance |
|---|---|---|---|---|
| CH₂/CH₃ Ratio | Significant increase | 6 hours | r² = 0.965-0.9868 | Membrane lipid alterations |
| Choline Resonance | Maintained at 3.2 ppm | Varies by inducer | Distinguishes from necrosis | Membrane integrity preservation |
| sn-Glycero-3-Phosphocholine (GPC) | Elevated levels | 3-4 days | Marker for senescence-associated apoptosis | Altered phospholipid metabolism |
| O-Phosphocholine (PC) | Decreased levels | 3-4 days | Marker for senescence | Phospholipid metabolism shift |
Distinctive NMR Profiles for Apoptosis Versus Necrosis
NMR spectroscopy effectively differentiates apoptosis from necrosis through distinct metabolic fingerprints. While apoptosis shows increased CH₂/CH₃ ratio with preserved choline resonance, necrosis exhibits markedly different patterns. In cadmium-induced apoptosis versus mercury-induced necrosis in MRC-5 normal human lung cells, the CH₂/CH₃ ratio increased only slightly within 2 hours of mercury exposure, with no further increase observed with longer exposure times [61]. Most notably, the choline resonance was abolished following 2-hour mercury exposure, a phenomenon absent in cadmium-induced apoptosis [61].
Research in senescent cancer cells, which share some features with apoptotic cells, reveals additional metabolic fingerprints. Senescent murine liver carcinoma H-Ras cells show elevated levels of sn-glycero-3-phosphocholine (GPC), myoinositol, taurine, and creatine, with concurrent decreases in glycine, O-phosphocholine (PC), threonine, and valine [62]. These metabolic alterations strongly impact growth and redox metabolism, revealing potential MRS signals for detecting senescent cancer cells in vivo [62].
Table 2: Comparative ¹H-NMR Spectral Profiles for Apoptosis vs. Necrosis
| Spectral Feature | Apoptosis | Necrosis | Discriminatory Power |
|---|---|---|---|
| CH₂/CH₃ Ratio | Significantly increased | Minimally increased | High (distinct pattern) |
| Choline Resonance | Maintained at 3.2 ppm | Abolished | High (definitive marker) |
| Mobile Lipid Levels | Gradual accumulation | Rapid changes | Moderate |
| Metabolic Profile | Ordered, specific changes | Disordered, nonspecific changes | High |
| Detection Window | Early (6+ hours) | Variable | Moderate |
Experimental Protocols for NMR-Based Apoptosis Detection
Cell Culture and Apoptosis Induction
Standardized protocols for NMR-based apoptosis detection begin with appropriate cell culture and reproducible apoptosis induction methods. For HeLa cells (human cervical carcinoma cells), researchers typically culture cells as monolayers in Dulbecco's Modified Eagle's Medium (DMEM) under 5% CO₂ at 37°C [7]. Apoptosis induction employs specific chemical inducers:
- Etoposide Treatment: The topoisomerase II inhibitor etoposide effectively induces apoptosis in HeLa cells [60]. As a DNA-damaging agent, etoposide triggers the intrinsic apoptotic pathway through mitochondrial membrane permeabilization.
- Doxorubicin Treatment: For experimental setups, doxorubicin added to culture medium at a final concentration of 5 μmol/L in a total volume of 1.5 mL reliably induces apoptosis [7]. This anthracycline chemotherapeutic agent intercalates into cellular DNA and inhibits topoisomerase II, causing double-strand breaks and activating p53-mediated apoptosis pathways.
- Cadmium Treatment: In MRC-5 normal human lung cells, cadmium effectively induces apoptosis as confirmed by phosphatidylserine externalization measured via flow cytometry [61].
For necrosis induction, researchers commonly use:
- Ethanol Treatment: High concentrations (99%) of ethanol induce nonspecific and rapid cellular damage resulting in necrosis [7]. Ethanol's lipophilic nature disrupts membrane integrity, promotes cellular leakage, and denatures proteins.
- Mercury Treatment: In MRC-5 cells, mercury exposure induces necrotic cell death [61].
- Other Necrosis Inducers: Ethacrynic acid or cytochalasin B also effectively induce necrosis in experimental systems [60].
Sample Preparation and NMR Spectroscopy Parameters
Proper sample preparation is critical for reproducible NMR-based apoptosis detection:
- Cell Harvesting: Collect cells at appropriate time points post-induction using trypsinization or mechanical scraping [62].
- Metabolite Extraction: Employ dual-phase extraction methods for polar metabolites. Add ice-cold methanol to cell pellets, vigorously vortex, then combine samples from multiple culture flasks [62]. Following addition of methyl tert-butyl ether (MTBE), sonicate samples using an UltraSonic Heatsystem [62].
- NMR Analysis: Conduct high-resolution ¹H-NMR spectroscopy on extracted metabolites. Studies frequently use 600 MHz systems for optimal resolution [62]. For live cell monitoring, magnetic resonance spectroscopy (MRS)-compatible cell perfusion systems enable dynamic metabolic tracking, with cells sometimes grown in three-dimensional conformation on microcarrier beads [62].
Data Processing and Validation Methods
NMR data processing and validation ensure accurate apoptosis detection:
- Spectral Analysis: Focus on specific spectral regions including methylene resonances (1.3 ppm), methyl resonances (0.9 ppm), and choline resonances (3.2 ppm) [60] [61].
- Ratio Calculation: Compute CH₂/CH₃ ratios by comparing integrated peak areas or intensities.
- Validation Techniques: Correlate NMR findings with established apoptosis assays:
NMR in Context: Comparative Analysis with Other Cell Death Detection Methods
Technical Comparisons with Morphological and Biochemical Assays
NMR spectroscopy provides distinct advantages and limitations compared to conventional apoptosis detection methods:
Table 3: Comparison of Apoptosis Detection Methods
| Method | Principle | Advantages | Limitations | Suitable Applications |
|---|---|---|---|---|
| ¹H-NMR Spectroscopy | Detection of metabolic fingerprints and intensity ratios | Label-free, non-destructive, quantitative, early detection, provides metabolic information | Lower sensitivity than specialized methods, requires specialized equipment, complex data analysis | Drug screening, mechanistic studies, in vivo translation |
| Flow Cytometry | Annexin V/PI staining for membrane changes | High throughput, quantitative, multi-parameter | Requires staining, cell destruction, limited structural information | High-throughput screening, immunology research |
| Fluorescence Microscopy | Fluorescent probes for apoptotic markers | Visual confirmation, subcellular localization | Photobleaching, potential dye toxicity, semi-quantitative | Cell biology studies, mechanism validation |
| Full-Field OCT | High-resolution interferometric imaging | Label-free, high-resolution 3D morphology, real-time monitoring | Limited molecular specificity, specialized equipment | Morphological assessment, single-cell analysis |
| Immunoblotting | Detection of apoptotic proteins (caspases) | Specific, sensitive, well-established | Destructive, semi-quantitative, no live cell monitoring | Mechanism confirmation, pathway analysis |
Integration with Morphological Assessment Techniques
Advanced morphological techniques like full-field optical coherence tomography (FF-OCT) complement NMR findings by providing high-resolution, label-free visualization of apoptotic morphological changes. FF-OCT captures characteristic apoptotic features including echinoid spine formation, cell contraction, membrane blebbing, and filopodia reorganization [7]. These morphological events typically manifest after the metabolic changes detectable by NMR, establishing a temporal relationship where NMR serves as an early detection method while imaging confirms subsequent structural alterations.
The integration of NMR metabolic fingerprinting with morphological assessment creates a comprehensive analytical platform for cell death research. NMR identifies early biochemical events in apoptosis through specific metabolic ratios, while imaging techniques validate the resulting phenotypic manifestations, together providing robust confirmation of cell death mechanisms.
Research Reagent Solutions for NMR-Based Apoptosis Studies
Table 4: Essential Research Reagents for NMR-Based Apoptosis Detection
| Reagent/Cell Line | Specification | Research Function | Application Context |
|---|---|---|---|
| HeLa Cells | Human cervical carcinoma cells (KCLB-10002) | Standardized apoptosis model system | Drug response studies, mechanism investigation |
| MRC-5 Cells | Normal human lung cell line | Non-cancerous apoptosis model | Toxicity testing, comparative studies |
| H-Ras Cells | Murine liver carcinoma cells | Senescence-associated apoptosis model | Cancer metabolism studies |
| Etoposide | Topoisomerase II inhibitor | Apoptosis inducer (DNA damage pathway) | Standardized apoptosis induction |
| Doxorubicin | Anthracycline chemotherapeutic | Apoptosis inducer (p53 pathway) | Chemotherapy response studies |
| Cadmium | Heavy metal solution | Apoptosis inducer in normal cells | Environmental toxin research |
| Dulbecco's Modified Eagle's Medium (DMEM) | With 10% fetal bovine serum | Standard cell culture maintenance | Consistent cell growth conditions |
| Methanol & MTBE | HPLC/spectroscopy grade | Metabolite extraction from cells | Sample preparation for NMR |
NMR spectroscopy establishes itself as a powerful analytical platform for biochemical fingerprinting of apoptosis, providing researchers with quantitative, non-destructive metabolic signatures that reliably distinguish programmed cell death from necrosis. The consistent increase in CH₂/CH₃ ratio intensity, coupled with preserved choline resonance, serves as a robust biomarker for apoptosis across multiple cell models. These metabolic fingerprints enable earlier detection of apoptosis compared to morphological methods, offering significant advantages for drug development and toxicology screening.
The integration of NMR metabolic data with complementary morphological techniques like FF-OCT creates a comprehensive framework for cell death analysis, connecting early biochemical events with subsequent structural manifestations. As research continues to refine NMR applications in cell death characterization, this technology promises to enhance our understanding of apoptotic pathways and improve assessment of therapeutic efficacy in biomedical research and drug development.
The precise differentiation between apoptosis and necrosis is a cornerstone of biomedical research, with significant implications for understanding disease pathogenesis and developing therapeutic strategies. While morphological features observed under a microscope provide the initial classification, the underlying biochemical pathways offer definitive mechanistic confirmation. This guide focuses on two cornerstone biochemical hallmarks of apoptosis: caspase activation and DNA laddering. We will objectively compare the experimental data and methodologies used to detect these markers, placing them within the critical context of validating morphological observations. The integration of these analytical layers provides researchers and drug development professionals with a robust framework for the accurate identification of programmed cell death.
Morphological Hallmarks: The Initial Diagnostic Landscape
The journey to classify cell death begins with visualizing cellular changes. High-resolution, label-free imaging techniques, such as Full-Field Optical Coherence Tomography (FF-OCT), have been instrumental in defining the distinct morphological landscapes of apoptosis and necrosis [7].
The table below summarizes the key morphological features that serve as the initial diagnostic criteria for distinguishing these two cell death pathways.
Table 1: Characteristic Morphological Features of Apoptosis and Necrosis
| Morphological Feature | Apoptosis | Necrosis |
|---|---|---|
| Cell Size | Cell contraction and shrinkage | Cell and organelle swelling (oncosis) |
| Plasma Membrane | Blebbing, integrity maintained until late stages | Rapid rupture and loss of integrity |
| Nucleus | Chromatin condensation, nuclear fragmentation | Karyolysis (nuclear dissolution) |
| DNA State | Condensed and marginalized chromatin | Dispersed chromatin |
| Inflammatory Response | None; silent phagocytosis | Significant; release of pro-inflammatory contents |
| Adhesion Structures | Reorganization and detachment | Abrupt loss of adhesion |
These morphological patterns, while highly indicative, are not entirely specific. For instance, certain stages of apoptosis can share features with necrosis, and some regulated forms of necrosis may exhibit overlapping morphology. Therefore, biochemical analysis is essential for unambiguous identification [63].
Biochemical Pathways: The Molecular Machinery of Apoptosis
At the biochemical level, apoptosis is executed by a family of cysteine proteases known as caspases. These enzymes exist as inactive zymogens and are activated through proteolytic cleavage in a cascade, leading to the controlled dismantling of the cell [64] [63].
The Caspase Cascade
Caspases are broadly categorized into initiator caspases (e.g., caspase-8, -9, -10) and effector caspases (e.g., caspase-3, -6, -7). Initiator caspases are activated in response to specific death signals, which trigger their aggregation and auto-activation within large multiprotein complexes such as the FADDosome (for extrinsic apoptosis) or the apoptosome (for intrinsic apoptosis) [64]. Once active, initiator caspases proteolytically activate effector caspases, which then cleave a wide array of cellular substrates, resulting in the characteristic morphological changes [63].
Key Apoptotic Substrates: From Caspases to DNA Laddering
Two of the most critical downstream events in apoptosis are:
- Cleavage of the DNA-repair enzyme PARP1 by effector caspases like caspase-3. This inactivates DNA repair mechanisms, facilitating the fragmentation of DNA [65].
- Activation of specific DNases, such as Caspase-Activated DNase (CAD). CAD is cleaved from its inhibitor (ICAD) by caspase-3, freeing it to cleave genomic DNA at internucleosomal regions, producing the classic DNA ladder—fragments in multiples of approximately 180 base pairs [63].
The following diagram illustrates the core biochemical pathway of caspase-dependent apoptosis.
Experimental Data: Quantitative Correlations
Correlating morphological observations with quantitative biochemical data is the definitive step for confirming the mechanism of cell death. The following table consolidates key experimental findings that link these analytical layers.
Table 2: Correlation of Morphological and Biochemical Apoptosis Markers
| Experimental Readout | Quantitative/Semi-Quantitative Data | Correlation with Morphology | Experimental Method |
|---|---|---|---|
| Caspase-3/7 Activity | Increase in enzymatic activity or cleavage products. | Coincides with appearance of membrane blebbing and cell shrinkage [7]. | Fluorochrome-labeled inhibitors (FLICA), immunoblotting for cleaved caspase-3 [66]. |
| PARP1 Cleavage | Shift from 116 kDa full-length to 89 kDa cleaved fragment. | Occurs upstream of definitive apoptotic body formation [65]. | Single-cell Western blot, standard immunoblotting [65]. |
| DNA Laddering | Discrete ~180 bp DNA fragments on gel electrophoresis. | A late event, associated with nuclear condensation and fragmentation [63]. | Agarose gel electrophoresis, Single-Cell Electrophoresis (Comet/SEVAP assay) [65]. |
| Membrane Integrity | Retention of viability dyes (early apoptosis) vs. uptake (necrosis/late apoptosis). | Apoptotic cells (Caspase-3+) are dye-excluding; necrotic cells are dye-permeable [67]. | Flow cytometry with Annexin V/PI or Cisplatin-based viability stains [67]. |
Advanced single-cell analysis, such as the Single-cell Electrophoresis-based Viability and Protein (SEVAP) assay, allows for the simultaneous measurement of multiple apoptotic events—DNA fragmentation and PARP1 cleavage—from the same cell. This powerful approach directly links biochemical pathways within a single morphological context, resolving heterogeneity in cell death responses [65].
Experimental Protocols: A Guide to Key Methodologies
Protocol 1: Single-Cell Analysis of Apoptosis via SEVAP Assay
The SEVAP assay is a high-throughput, multimodal endpoint assay designed to elucidate cell-to-cell heterogeneity in apoptosis signaling [65].
- Cell Isolation and Lysis: Isolate single cells in microwell arrays molded in a thin polyacrylamide gel (PAG). Lyse cells directly in the wells using a buffer containing SDS and Triton X-100 to denature proteins and release cellular contents [65].
- Electrophoretic Separation: Perform polyacrylamide gel electrophoresis (PAGE) to separate proteins and DNA based on molecular weight. This step baseline-resolves intact DNA from fragmented DNA, and full-length PARP1 (116 kDa) from its cleaved fragment (89 kDa) [65].
- Photo-immobilization: Expose the gel to UV light to activate benzophenone cross-linkers covalently incorporated into the PAG, permanently immobilizing the separated proteins within the gel matrix [65].
- Multimodal Staining and Detection:
- DNA Staining: Stain double-stranded DNA with a fluorescent intercalating dye like SYBR Green I to visualize the DNA "comet" or laddering pattern for each cell.
- Protein Immunoprobing: Probe the immobilized proteins with primary antibodies (e.g., anti-PARP1, anti-β-tubulin) followed by fluorescently labeled secondary antibodies [65].
- Imaging and Analysis: Acquire fluorescence images. Analyze the electrophoretic mobility of DNA and PARP1 for each single cell to classify its apoptotic state (non-apoptotic vs. apoptotic) [65].
Protocol 2: Distinguishing Apoptosis and Necrosis via Flow Cytometry with Viability Stains
This protocol leverages the key morphological hallmark of membrane integrity to distinguish early apoptotic cells from necrotic cells.
- Cell Staining: Resuspend cells in a binding buffer and stain with:
- Annexin V conjugated to a fluorochrome: Binds to phosphatidylserine (PS) exposed on the outer leaflet of the plasma membrane in apoptotic cells.
- A membrane-impermeant viability dye such as Propidium Iodide (PI) or Cisplatin: Enters cells only when membrane integrity is lost, marking necrotic or late apoptotic cells [67].
- Incubation and Analysis: Incubate cells for 10-20 minutes at room temperature and analyze immediately by flow cytometry.
- Data Interpretation:
- Annexin V⁻ / Viability Dye⁻: Viable, non-apoptotic cells.
- Annexin V⁺ / Viability Dye⁻: Early apoptotic cells (intact membrane).
- Annexin V⁺ / Viability Dye⁺: Late apoptotic or necroptotic cells (compromised membrane).
- Annexin V⁻ / Viability Dye⁺: Necrotic cells or cellular debris [67].
This workflow is summarized in the diagram below.
The Scientist's Toolkit: Essential Reagents and Solutions
Successful experimentation in cell death research relies on a suite of specific reagents and tools. The following table details key solutions used in the featured experiments.
Table 3: Key Research Reagent Solutions for Apoptosis Analysis
| Research Tool | Function / Target | Application in Experiments |
|---|---|---|
| FLICA (Fluorochrome-Labeled Inhibitors of Caspases) | Irreversibly binds to active caspase enzymes. | Flow cytometric or fluorescent microscopic detection of specific caspase (e.g., 8, 9) activity in live cells [66]. |
| Antibody: Cleaved Caspase-3 | Recognizes the activated, cleaved form of caspase-3. | Immunoblotting, flow cytometry, and immunohistochemistry to confirm executioner caspase activation [67]. |
| Antibody: Cleaved PARP1 | Distinguishes the 89 kDa apoptotic fragment from full-length PARP1. | Key biomarker for apoptosis in immunoblotting and single-cell Western blot assays (e.g., SEVAP) [65]. |
| SYBR Green I / Propidium Iodide | Fluorescent nucleic acid stains. | SYBR Green I: stains DNA in fixed/permeabilized assays (e.g., SEVAP). PI: viability dye for flow cytometry [65]. |
| Annexin V Conjugates | Binds to externalized Phosphatidylserine (PS). | Flow cytometric identification of early apoptotic cells when used with a viability dye [63]. |
| Cisplatin-based Viability Stain | DNA-binding agent that only enters cells with compromised membranes. | Used as a viability dye in mass cytometry (CyTOF) and flow cytometry to identify necrotic cells [67]. |
The unequivocal classification of cell death requires a multi-faceted approach that rigorously correlates distinct morphological features with definitive biochemical markers. While visualization of cell shrinkage, blebbing, and nuclear condensation provides the initial evidence for apoptosis, confirmation hinges on demonstrating the activation of the caspase cascade and its downstream effects, such as PARP1 cleavage and DNA laddering. Modern techniques, including single-cell multimodal assays like SEVAP and advanced flow cytometry, now allow for an unprecedented level of integration between these morphological and biochemical datasets. This rigorous, correlated framework is essential for researchers and drug developers to accurately assess mechanisms of cell death in disease models and therapy development.
In the fields of cell biology and drug development, accurately distinguishing between apoptosis and necrosis is fundamental to understanding cellular responses to toxins, therapies, and disease processes. While numerous biochemical assays exist, morphological assessment remains the gold standard for definitively classifying cell death pathways, providing an irreplaceable layer of biological context [6]. The core distinction lies in the pattern of death: apoptosis is a highly regulated, energy-dependent process characterized by cell shrinkage, membrane blebbing, and formation of apoptotic bodies that are neatly phagocytosed, whereas necrosis represents a catastrophic failure of membrane integrity leading to cell swelling, organelle breakdown, and inflammatory spillage of contents [3] [6]. This guide provides a structured, practical workflow for selecting the optimal detection assay based on your specific research question, with a firm grounding in the morphological hallmarks that define each death pathway.
Core Morphological and Biochemical Hallmarks
A confident diagnosis of cell death type relies on recognizing a constellation of specific morphological and biochemical features. The table below summarizes the defining characteristics of apoptosis and necrosis, which form the basis for most detection assays.
Table 1: Key Morphological and Biochemical Hallmarks of Apoptosis vs. Necrosis
| Feature | Apoptosis | Necrosis |
|---|---|---|
| Overall Process | Programmed, regulated, energy-dependent [3] | Unregulated, passive, catastrophic [3] |
| Cell Morphology | Cell shrinkage, membrane blebbing, formation of apoptotic bodies [6] | Cell and organelle swelling, loss of membrane integrity [3] [6] |
| Nuclear Morphology | Chromatin condensation (pyknosis), nuclear fragmentation (karyorrhexis) [6] | Karyolysis (nuclear dissolution), less organized condensation [68] |
| Plasma Membrane | Integrity maintained until late stages; Phosphatidylserine (PS) externalization [69] | Rapid loss of integrity, rupture, and leakage of cellular contents [3] |
| Key Biochemical Markers | Caspase activation (especially caspase-3/7), internucleosomal DNA cleavage [70] [69] | Absence of significant caspase activation, random DNA digestion [35] |
| Inflammatory Response | None ("silent" removal) [68] | Significant; triggers inflammation [3] |
Assay Comparison: Principles, Applications, and Methodologies
A wide array of assays is available, each detecting different features from the hallmarks table. The choice of assay depends on the research question, the need for quantification, throughput, and whether live or fixed cells are used.
Table 2: Comparison of Key Apoptosis and Necrosis Detection Assays
| Assay Name | Principle / Target | Morphological Context | Throughput | Key Advantages | Key Limitations |
|---|---|---|---|---|---|
| Microscopy (Light/Electron) | Direct visualization of cell shrinkage, membrane blebbing, apoptotic bodies, nuclear condensation [6] | Direct | Low | Direct morphological assessment; considered a definitive standard [6] | Low throughput, subjective, requires expertise [7] |
| Annexin V / Propidium Iodide (PI) | Binds to externalized PS (early apoptosis) and PI stains DNA in membrane-compromised cells (necrosis/late apoptosis) [71] | Indirect | Medium | Distinguishes live, early apoptotic, and late apoptotic/necrotic populations [71] | Cannot distinguish late apoptosis from primary necrosis; PS exposure not exclusive to apoptosis [35] |
| Caspase Activity Assays | Measures cleavage of fluorogenic or luminogenic substrates (e.g., DEVD) by executioner caspases-3/7 [69] | Indirect | High (HTS compatible) | Highly specific for apoptosis; very sensitive and quantitative [69] | Misses caspase-independent cell death pathways [25] |
| TUNEL Assay | Labels DNA strand breaks (a late event in apoptosis) [70] | Indirect | Low | Specific for DNA fragmentation [70] | Can give false positives in necrotic cells with random DNA degradation [70] |
| Live-Cell Imaging (FRET/FF-OCT) | FRET: Visualizes caspase activation in real-time. FF-OCT: Label-free, high-resolution 3D morphology [35] [7] | Direct | Medium | Real-time kinetic data; distinguishes primary from secondary necrosis [35] | Requires specialized equipment and stable cell lines (FRET) [35] |
Detailed Experimental Protocols
To ensure experimental reproducibility, below are summarized protocols for two commonly used and one advanced assay.
Protocol 1: Annexin V/7-AAD Staining for Flow Cytometry (Adapted from ab176749 [71]) This protocol allows for the quantification of live, early apoptotic, and late apoptotic/necrotic cell populations.
- Cell Preparation: Harvest cells (suspension or trypsinized adherent cells) and wash with cold PBS.
- Resuspension: Resuspend cell pellet in 1X Annexin V Binding Buffer at a concentration of 1 x 10^6 cells/mL.
- Staining: Add Annexin V-FITC and 7-AAD (a viability dye similar to PI) to the cell suspension. Gently vortex and incubate for 15-20 minutes at room temperature in the dark.
- Analysis: Within 1 hour, analyze by flow cytometry. Use FITC (FL1) and 7-AAD (FL3) channels, collecting data from at least 10,000 events per sample.
Protocol 2: Caspase-3/7 Activity Luminescent Assay (Adapted from NCBI Assay Guidance Manual [69]) This is a homogeneous, high-throughput protocol for quantifying apoptosis.
- Plate Cells: Seed cells in an opaque-walled, white microplate and treat with compounds.
- Equilibrate Reagents: Equilibrate Caspase-Glo 3/7 reagent to room temperature.
- Add Reagent: Add an equal volume of Caspase-Glo 3/7 reagent to each well.
- Mix and Incubate: Mix contents on a plate shaker for 30 seconds and incubate at room temperature for 30-60 minutes to allow signal development.
- Read: Measure the luminescence using a plate-reading luminometer.
Protocol 3: Label-Free Morphological Assessment via FF-OCT (Adapted from [7]) This protocol uses Full-Field Optical Coherence Tomography for non-invasive, 3D live-cell imaging.
- Cell Preparation: Plate cells on imaging dishes and allow to adhere.
- Induction: Induce apoptosis (e.g., with 5 µM Doxorubicin) or necrosis (e.g., with high-concentration Ethanol).
- Image Acquisition: Place the dish on the FF-OCT system. Initiate time-lapse imaging immediately after induction, acquiring en face (x-y) cross-sectional images at regular intervals (e.g., every 20 minutes) for up to 3 hours.
- 3D Reconstruction: Stack the acquired tomographic images to reconstruct the 3D volume and surface morphology of the cells.
- Analysis: Identify apoptotic cells by characteristic echinoid spine formation, contraction, and membrane blebbing. Identify necrotic cells by rapid membrane rupture and loss of adhesion structure.
Integrated Workflow and Pathway Diagrams
A strategic workflow is essential for accurate cell death classification. The following diagram and logic map guide assay selection based on the research question and available resources.
Diagram 1: Assay Selection Workflow
The relationship between the key molecular events in apoptosis and necrosis and the assays that detect them is complex. The following pathway diagram visualizes these connections, highlighting critical decision points for assay selection.
Diagram 2: Cell Death Pathways and Assay Targets
The Scientist's Toolkit: Essential Research Reagent Solutions
The following table catalogs key reagents and their functions, forming the essential toolkit for conducting experiments in this field.
Table 3: Key Research Reagent Solutions for Apoptosis/Necrosis Detection
| Reagent / Kit | Function / Target | Key Feature | Example Application |
|---|---|---|---|
| Caspase-Glo 3/7 Assay [69] | Luminescent measurement of caspase-3/7 activity via cleavage of DEVD-aminoluciferin. | Homogeneous "add-mix-measure" format; highly sensitive and HTS-compatible. | High-throughput screening of compound libraries for pro-apoptotic agents. |
| Annexin V-FITC / 7-AAD Kit [71] | Fluorescent conjugates to detect PS exposure (Annexin V) and membrane integrity (7-AAD). | Allows multiplexed distinction of live, early apoptotic, and late apoptotic/necrotic cells. | Flow cytometry analysis to determine the mode of cell death induced by a new drug candidate. |
| FAM-FLICA & 7-AAD Kit [72] | Fluorescent-labeled caspase inhibitor (FAM-FLICA) labels active caspases; 7-AAD labels dead cells. | Simultaneously detects caspase activity (apoptosis) and membrane integrity (necrosis) in a single sample. | Detailed flow cytometric analysis of cytotoxicity from immunologic agents (e.g., T cells). |
| Hoechst 33342 / DAPI [6] | Cell-permeable and impermeable DNA stains, respectively. | Visualize nuclear morphology (condensation, fragmentation) by fluorescence microscopy. | Qualitative and quantitative assessment of apoptotic nuclei in fixed or live (Hoechst) cells. |
| Genetic Biosensors (e.g., FRET-DEVD) [35] | Genetically encoded probe that loses FRET upon caspase-mediated cleavage. | Enables real-time, single-cell kinetic analysis of caspase activation in live cells. | Studying the dynamics and heterogeneity of apoptosis initiation in a cell population. |
Selecting the right assay for cell death research is not a one-size-fits-all process. A practical workflow begins with a clear research objective and leverages the strengths of different methodologies. For high-throughput screening, caspase activity assays are unparalleled. For distinguishing early and late stages of death in a heterogeneous population, Annexin V/7-AAD staining by flow cytometry is the workhorse. However, when results from these indirect assays are ambiguous, or when definitive classification is required, morphological assessment by microscopy remains the indispensable arbitrator [6]. The future of the field lies in the adoption of advanced, label-free technologies like FF-OCT that provide high-resolution 3D morphological data in real-time, bridging the gap between the simplicity of biochemical assays and the rich biological context of traditional pathology [7]. By grounding your experimental strategy in the foundational principles of cell morphology, you can ensure accurate, reliable, and interpretable results in your research on apoptosis and necrosis.
Solving the Puzzle: Overcoming Common Challenges in Differentiating Cell Death Pathways
In cell death research, accurately distinguishing between apoptosis and necrosis is fundamental, yet the phenomenon of secondary necrosis presents a significant diagnostic challenge. Secondary necrosis describes the natural outcome of the complete apoptotic program when apoptotic cells are not cleared by scavengers, leading to a terminal phase that morphologically resembles necrosis [73] [74]. This process is not a separate death pathway but rather the autolytic disintegration of cells that have already executed apoptosis. In vitro, where scavenger cells are typically absent, this progression is almost universal: apoptosing cells, after initiating self-dismantling with an initially intact membrane, ultimately swell, experience irreparable membrane damage, and disrupt, culminating in the release of intracellular contents [73]. In vivo, secondary necrosis occurs when massive apoptosis overwhelms the available scavenging capacity or when the scavenger mechanism is directly impaired, potentially leading to pathogenic inflammation and autoimmune responses due to the leakage of cell contents [73]. For researchers and drug development professionals, misclassifying these late-apoptotic cells as primary necrotic cells can lead to a fundamental misinterpretation of experimental outcomes, toxicological assessments, and therapeutic efficacy.
Comparative Morphological and Biochemical Hallmarks
The following tables summarize the key characteristics that differentiate the various cell death states, providing a reference for accurate identification.
Table 1: Morphological and Biochemical Changes Across Cell Death Modes
| Feature | Apoptosis (Execution Phase) | Secondary Necrosis | Primary Necrosis |
|---|---|---|---|
| Cell Shape & Volume | Cell rounding, shrinkage, and blebbing [73] [75] | Swelling of the cell or apoptotic bodies [73] | Cytoplasmic and mitochondrial swelling [73] [76] |
| Plasma Membrane | Near-to-intact; blebbing with intact integrity [73] [75] | Generalized, irreparable damage and rupture [73] | Loss of integrity and increased permeability; rapid rupture [75] |
| Nuclear Changes | Nuclear fragmentation, intense chromatin condensation (pyknosis) [73] [75] | Nuclear fragmentation and intense chromatin condensation [73] | Moderate chromatin condensation; no nuclear fragmentation [73] |
| Organelles | Mitochondrial "thread-grain" transition; organelles still functional [73] [76] | Intense mitochondrial swelling [73] | Organelles swell, disintegrate, and become non-functional [73] [76] |
| DNA Fragmentation | Endonuclease-induced cleavage into a ladder pattern [75] | - | Random degradation of genomic DNA [75] |
| Inflammatory Response | Usually no inflammatory response [75] [77] | Induction of tissue injury and inflammatory/autoimmune responses [73] | Enhancement of inflammatory response [75] |
| Clearence Mechanism | Phagocytosis by scavengers (efferocytosis) [73] [75] | Cell disruption and lysis [73] [75] | Cell lysis [75] |
Table 2: Kinetic and Mechanistic Profiles of Necrotic Processes
| Parameter | Secondary Necrosis | TNF-Induced Necroptosis | H2O2-Induced Necrosis |
|---|---|---|---|
| Initiating Stimulus | Completion of apoptotic program without phagocytosis [73] | TNFα receptor engagement, especially when caspases are inhibited [75] [78] | Direct oxidative stress and physicochemical injury [78] |
| Signaling Phase | Preceded by full apoptotic execution phase (caspase activation, etc.) [73] | Defined signaling phase involving RIP1/RIP3/MLKL (necrosome) [75] [78] | No defined signaling phase; starts immediately with LMP [78] |
| Key Molecular Dependencies | Caspases, self-hydrolytic enzymes [73] | RIP1 kinase, mitochondrial complex I, cPLA2 [78] | Iron-dependent Fenton reactions [78] |
| Kinetics of Disintegration | Slower, follows apoptotic phase [73] | Regulated, with a defined sequence of subcellular events [78] | Rapid, direct damage [78] |
| Sequence of Terminal Events | Swelling → membrane damage → disruption [73] | Oxidative burst → MTP hyperpolarization → LMP → PMP [78] | LMP → oxidative burst → MTP hyperpolarization → PMP [78] |
| LMP: Lysosomal Membrane Permeabilization; PMP: Plasma Membrane Permeabilization; MTP: Mitochondrial Transmembrane Potential |
Experimental Protocols for Differentiation
High-Resolution, Label-Free Imaging of Single Cells
Application: This protocol uses Full-Field Optical Coherence Tomography (FF-OCT) to distinguish apoptosis from necrosis in real-time based on morphology without labels, preserving native cell state [7].
Workflow Diagram: Label-Free Cell Death Imaging
Detailed Procedure:
- Cell Preparation: Culture HeLa cells (or other relevant cell line) as a monolayer in Dulbecco's Modified Eagle's Medium (DMEM) under standard conditions (5% CO₂, 37°C) [7].
- Death Induction:
- For Apoptosis: Add doxorubicin to the culture medium at a final concentration of 5 μmol/L. Doxorubicin intercalates into DNA, causing double-strand breaks and activating p53-mediated apoptotic pathways [7].
- For Necrosis: Treat cells with 99% ethanol. Ethanol acts as a lipophilic agent that disrupts the phospholipid bilayer and denatures proteins, leading to rapid loss of membrane integrity [7].
- FF-OCT Imaging: Use a custom-built time-domain FF-OCT system with a broadband halogen light source (e.g., center wavelength 650 nm) and a Linnik-configured interferometer with high numerical aperture (NA: 0.8) objectives. This setup achieves sub-micrometer resolution [7].
- Data Acquisition: Initiate imaging immediately after drug administration. Acquire en face (x-y) cross-sectional images continuously at 20-minute intervals for up to 180 minutes. Use a piezoelectric actuator in the reference arm for phase-shifting to isolate sample reflection information [7].
- 3D Reconstruction & Analysis: Stack the tomographic images to reconstruct 3D surface topography. Quantify morphological changes.
- Key Apoptotic Features to Quantify: Cell contraction, echinoid spine formation on the membrane, membrane blebbing, and filopodia reorganization [7].
- Key Necrotic Features to Quantify: The timing and extent of membrane rupture, leakage of intracellular contents, and abrupt loss of adhesion structures [7].
Multiparametric Fluorescence Assay for Kinetic Profiling
Application: This protocol uses a combination of fluorescent probes and high-resolution time-lapse microscopy to kinetically distinguish secondary necrosis from other death modes based on a sequence of subcellular events [78].
Workflow Diagram: Kinetic Profiling of Cell Death
Detailed Procedure:
- Cell Preparation: Seed appropriate cells (e.g., mouse embryonic fibroblasts, L929 cells) in imaging-compatible plates. If studying specific pathways, transfect with relevant constructs (e.g., optogenetic switches like OptoBAX for precise apoptosis initiation) [79] [78].
- Fluorescent Staining: Load cells with a panel of fluorescent probes:
- CM-H2DCFDA: For detecting reactive oxygen species (ROS) and oxidative burst [78].
- TMRM (Tetramethyl rhodamine methyl ester): For monitoring mitochondrial membrane potential (hyperpolarization or loss) [78].
- Acridine Orange: For assessing lysosomal membrane permeabilization (LMP). A loss of red acridine orange fluorescence indicates LMP [78].
- Propidium Iodide (PI): A membrane-impermeant dye that enters cells only upon plasma membrane permeabilization (PMP), marking the terminal step [73] [78].
- Death Induction & Imaging: Apply the death stimulus (e.g., TNFα + caspase inhibitor for necroptosis, anti-Fas for secondary necrosis, H₂O₂ for primary necrosis). Immediately begin high-resolution time-lapse microscopy to track all fluorescent channels over time [78].
- Kinetic Analysis: Analyze the sequence and timing of the subcellular events.
- Secondary Necrosis (post-apoptosis): Preceded by apoptotic markers (caspase activation, phosphatidylserine exposure). The disintegration phase shows the sequence: oxidative burst → mitochondrial membrane hyperpolarization → LMP → PMP [78].
- Necroptosis: Initiated by a defined signaling phase (RIP1/RIP3/MLKL), followed by the same terminal sequence of events as secondary necrosis, though with different kinetics [78].
- Primary Necrosis (H₂O₂-induced): Starts immediately with LMP, followed by oxidative burst, mitochondrial hyperpolarization, and PMP [78].
Key Signaling Pathways and Molecular Mechanisms
Understanding the molecular regulators is crucial for designing experiments that can definitively separate these processes.
Diagram: Core Apoptotic and Necroptotic Pathways
Pathway Key Insights:
- Apoptosis to Secondary Necrosis: The intrinsic apoptotic pathway is mediated by BCL-2 family proteins like BAX. Upon activation, BAX undergoes conformational change and inserts into the outer mitochondrial membrane, forming pores that lead to the release of cytochrome c, which activates the caspase cascade [75] [79]. The cell is then dismantled in a controlled manner. If not cleared by phagocytosis (efferocytosis), this leads to secondary necrosis, an autolytic process where the cell finally loses membrane integrity [73] [74]. A newly identified regulatory mechanism involves the mitochondrial protein VDAC1, which, under stress, can bind and deactivate the apoptotic inhibitor Bcl-xL, thereby promoting apoptosis and its potential progression to secondary necrosis [80].
- Necroptosis: This is a backup cell death pathway when apoptosis is blocked. Triggered by ligands like TNFα in the absence of caspase activity, it depends on the kinase activities of RIP1 and RIP3, which form a complex called the necrosome [75] [78]. The necrosome phosphorylates the effector MLKL, causing it to oligomerize and insert into the plasma membrane, directly causing membrane rupture and release of damage-associated molecular patterns (DAMPs) [75].
The Scientist's Toolkit: Essential Reagents and Solutions
Table 3: Key Research Reagents for Differentiating Cell Death Pathways
| Reagent / Tool | Primary Function | Application in Distinguishing Death Modes |
|---|---|---|
| Caspase-3 Antibody [75] | Detects activated executioner caspase-3 via IHC/IF. | Positive staining confirms apoptosis is underway, helping rule out primary necrosis or pure necroptosis. |
| BAX Antibody [75] | Labels activated BAX protein during intrinsic apoptosis. | Identifies commitment to the mitochondrial apoptotic pathway, a precursor to secondary necrosis. |
| Propidium Iodide (PI) [73] [78] | Membrane-impermeant DNA dye. | Stains cells only upon loss of membrane integrity. Used to mark terminal stages (secondary/primary necrosis). Must be combined with early apoptotic markers. |
| Annexin V Conjugates | Binds externalized phosphatidylserine (PS). | Marks early/mid-stage apoptosis when membrane is intact (PI-negative). Late-stage secondary necrosis is Annexin V+/PI+. |
| Necrostatin-1 [78] | Specific inhibitor of RIP1 kinase. | Used to functionally test for necroptosis. Inhibition of death in the presence of a caspase inhibitor confirms necroptotic pathway. |
| Z-VAD-FMK (pan-caspase inhibitor) | Irreversibly inhibits caspase activity. | Blocks apoptosis. Can be used to shift cell fate to necroptosis (if molecular machinery is present), aiding pathway dissection. |
| OptoBAX System [79] | Optogenetic switch for light-activated BAX oligomerization. | Precisely and non-invasively initiates intrinsic apoptosis on demand, allowing detailed study of its progression to secondary necrosis. |
| Acridine Orange [78] | Metachromatic dye for lysosomes. | Loss of red fluorescence indicates lysosomal membrane permeabilization (LMP), a key event in the disintegration phase of various necrotic deaths. |
| RIP3 / MLKL Antibodies [75] | Detect key necroptosis signaling proteins (IHC, IF, IP). | Phospho-specific antibodies confirm activation of the necroptotic pathway, distinguishing it from other necrotic processes. |
Misinterpreting secondary necrosis remains a significant pitfall in cell death research, but it can be avoided with a multifaceted experimental strategy. The key is to move beyond single-parameter assays. Best practices include: i) employing multiparametric, kinetic analyses using live-cell imaging to track the sequence of events; ii) combining functional tests (e.g., caspase inhibitors, necrostatins) with morphological and biochemical markers; and iii) utilizing high-resolution, label-free imaging technologies like FF-OCT to observe pristine morphological changes. By integrating these approaches and understanding the distinct molecular pathways, researchers can accurately classify cell death modes, thereby generating more reliable data for drug discovery and fundamental biological research.
Within the broader thesis on the specificity of morphological features for apoptosis versus necrosis research, a fundamental challenge emerges: accurately differentiating between two necrotic-looking cell death modalities—accidental necrosis and necroptosis. Historically, all necrotic cell death was considered an unprogrammed, passive process resulting from overwhelming chemical or physical insult [81]. This view has been revolutionized by the discovery of necroptosis, a form of programmed necrosis that is genetically encoded and tightly regulated [40] [34]. While both pathways culminate in plasma membrane rupture and release of intracellular contents, their origins, regulatory mechanisms, and implications for health and disease are profoundly different.
Failure to distinguish between these processes represents a significant pitfall in experimental research. Accidental necrosis is a passive consequence of cellular trauma, whereas necroptosis is an active signaling cascade that can be inhibited pharmacologically or genetically [82] [83]. This distinction is not merely semantic; it has direct consequences for understanding disease pathogenesis and developing targeted therapies. For researchers, scientists, and drug development professionals, employing precise discriminatory methodologies is therefore paramount for accurate experimental interpretation in the study of cell death.
Comparative Analysis: Core Characteristics
The following table summarizes the key differential features between accidental necrosis and necroptosis, providing a foundational reference for their identification.
Table 1: Fundamental characteristics of accidental necrosis versus necroptosis
| Characteristic | Accidental Necrosis | Necroptosis |
|---|---|---|
| Regulation | Unprogrammed, passive, and uncontrolled [81] | Programmed, active, and tightly regulated [81] [40] |
| Inducing Factors | Extreme physical/chemical stress, ischemia, toxins, mechanical trauma [82] | Death receptor ligands (e.g., TNF-α), pathogen sensors, caspase inhibition [81] [34] |
| Key Molecular Mediators | None (lack of specific molecular executors) | RIPK1, RIPK3, MLKL (core signaling pathway) [81] [40] |
| Energy Requirement | ATP-independent | ATP-dependent [82] |
| Inhibitors | Not inhibitable by specific agents | Necrostatin-1 (RIPK1 inhibitor), specific MLKL inhibitors [81] [84] |
| Immunogenicity | Highly inflammatory due to uncontrolled DAMP release [81] | Highly immunogenic; DAMP release can be tailored to the trigger [83] [34] |
Morphological and Biochemical Hallmarks
Despite a similar terminal phenotype of membrane rupture, careful observation can reveal differences in the progression of accidental necrosis and necroptosis. The table below details the distinct morphological and biochemical markers that facilitate their discrimination.
Table 2: Key markers for differentiating accidental necrosis from necroptosis
| Feature | Accidental Necrosis | Necroptosis |
|---|---|---|
| Cell Swelling | Rapid, high-amplitude swelling leading to swift rupture [85] | Cell swelling is a key feature, progressing to membrane rupture [81] |
| Organelle Integrity | Swelling and rupture of mitochondria and lysosomes [82] | Swelling of organelles, followed by disintegration of the cytoplasm and nucleus [81] |
| Plasma Membrane | Rapid loss of integrity, uncontrolled release of contents [85] | MLKL-mediated pore formation, leading to permeabilization and rupture [81] [40] |
| Caspase Dependence | Caspase-independent | Caspase-independent; often occurs when caspases are inhibited [81] [84] |
| Key Molecular Markers | Lack of specific phosphorylation markers | Phosphorylation of RIPK1, RIPK3, and MLKL [81] |
| Specific Inhibition | Not applicable | Inhibitable by Necrostatin-1, RIPK3 knockout, or MLKL deficiency [81] [34] |
The Necroptotic Signaling Pathway
The molecular pathway of necroptosis is well-defined and centers on a core signaling cascade. It is often initiated when death receptors (like TNFR1) are stimulated by their ligands (e.g., TNF-α) under conditions where caspase-8 activity is suppressed [40] [34]. This leads to the deubiquitination of RIPK1, which then interacts with RIPK3 via their RHIM domains to form a structure called the necrosome [40]. Within this complex, RIPK3 phosphorylates the terminal effector, MLKL. Phosphorylated MLKL undergoes oligomerization and translocates to the inner leaflet of the plasma membrane, where it integrates and forms pores [81] [83]. This pore formation leads to ion dysregulation, osmotic swelling, and the eventual rupture of the plasma membrane, resulting in the release of damage-associated molecular patterns (DAMPs) that drive inflammation [81] [34].
Experimental Protocols for Discrimination
A robust strategy to distinguish necroptosis from accidental necrosis involves a combination of pharmacological, genetic, and biochemical approaches. The following workflow provides a recommended protocol for conclusive identification.
Integrated Experimental Workflow
Detailed Methodologies for Key Experiments
1. Morphological Assessment by Microscopy
- Phase-Contrast Microscopy: Observe live cells for morphological changes. Necroptotic cells display early plasma membrane blebbing and progressive swelling, followed by rupture. This contrasts with the rapid, high-amplitude swelling seen in accidental necrosis [85].
- Nuclear Staining: Use fluorescent DNA-binding dyes (e.g., Hoechst 33342, DAPI) or perform eosin-methylene blue staining on cytospin preparations to analyze nuclear morphology. Unlike apoptosis, necroptosis does not typically feature pronounced nuclear fragmentation and condensation [85].
2. Flow Cytometry with Annexin V/Propidium Iodide (PI) This assay is crucial for tracking membrane integrity but requires careful interpretation.
- Protocol: Harvest cells and resuspend in binding buffer. Stain with Annexin V-FITC and PI according to manufacturer instructions. Analyze by flow cytometry within one hour [85].
- Interpretation: Both necroptotic and accidental necrotic cells will eventually become Annexin V and PI positive due to loss of membrane integrity. Kinetic analysis is key: in necroptosis, Annexin V positivity often precedes a gradual increase in PI uptake, reflecting the programmed pore formation. Accidental necrosis often shows a more simultaneous and rapid uptake of both dyes [85].
3. Inhibition of Necroptosis with Necrostatin-1
- Protocol: Pre-treat cells with Necrostatin-1 (e.g., 10-100 µM) for 30-60 minutes before applying the necroptotic stimulus (e.g., TNF-α plus a SMAC mimetic like SM-164 and a pan-caspase inhibitor like Z-VAD-FMK). Include vehicle control [81] [84].
- Assessment: Measure cell viability 4-24 hours later using assays like MTT, ATP-lite, or flow cytometry. A significant rescue of viability with Necrostatin-1 strongly indicates a necroptotic component. Accidental necrosis is not inhibited [81].
4. Detection of Pathway Phosphorylation
- Western Blotting: Lyse cells and perform immunoblotting with antibodies against key phosphorylated residues: phospho-RIPK1 (Ser166), phospho-RIPK3 (Ser227 in human), and phospho-MLKL (Ser358 in human) [81] [40]. The presence of these bands is a hallmark of active necroptotic signaling.
- Immunofluorescence: Fix and permeabilize cells. Stain with antibodies against phospho-MLKL. Activated MLKL shows translocation from the cytosol to punctate structures at the plasma membrane, providing visual confirmation of pathway activation [81].
The Scientist's Toolkit: Essential Reagents
The following table lists key reagents and tools essential for researching and discriminating necroptosis.
Table 3: Key research reagents for necroptosis studies
| Reagent / Tool | Function / Specificity | Example Use Case |
|---|---|---|
| Necrostatin-1 | A specific allosteric inhibitor of RIPK1 kinase activity [81] [84] | Pharmacological confirmation of RIPK1-dependent necroptosis in vitro and in vivo. |
| GSK'872 / NSA | Potent and selective inhibitors of RIPK3 kinase activity. | To confirm RIPK3's role downstream of RIPK1 or in RIPK1-independent pathways. |
| Phospho-Specific Antibodies | Detect active, phosphorylated forms of RIPK1 (pS166), RIPK3 (pS227), and MLKL (pS358) [81]. | Definitive biochemical verification of necrosome formation and MLKL activation via Western blot or IF. |
| Z-VAD-FMK | A broad-spectrum, cell-permeable caspase inhibitor. | To block apoptotic pathways and unmask necroptosis in experimental models [84]. |
| siRNA / shRNA / CRISPR-Cas9 | For genetic knockout or knockdown of RIPK1, RIPK3, or MLKL. | Provides definitive genetic evidence for the involvement of a specific protein in the cell death process. |
| Recombinant TNF-α + SM-164 + Z-VAD | A canonical combination to induce robust necroptosis; TNF activates signaling, SM-164 degrades cIAPs, Z-VAD blocks apoptosis [81]. | A positive control for triggering necroptosis in vitro. |
| Annexin V Detection Kits | Detect phosphatidylserine (PS) exposure on the outer leaflet of the plasma membrane. | Used in combination with PI to monitor the kinetics of membrane alterations during cell death [85]. |
The Role of Caspase Inhibition in Unmasking Alternative Death Pathways
The historical classification of cell death into the distinct categories of apoptosis and necrosis has been fundamentally challenged by the use of caspase inhibitors. These pharmacological tools have revealed that when the canonical apoptotic pathway is blocked, cells readily activate alternative backup death programs, including necroptosis and autophagic cell death. This review objectively compares the morphological and biochemical features of these cell death pathways, with a focus on data generated through caspase inhibition experiments. The findings underscore that caspase inhibition is not a panacea for cell death-related pathologies but rather a switch that can redirect cell fate towards alternative, and often inflammatory, demise.
Caspases, a family of cysteine-dependent proteases, are the principal executioners of programmed cell death (PCD) via apoptosis. The use of caspase inhibitors, such as the broad-spectrum compound zVAD-fmk, was initially envisioned as a primary therapeutic strategy to curb unwanted apoptosis in degenerative diseases and ischemia-reperfusion injuries [86] [87]. However, experimental data have consistently demonstrated that caspase inhibition often fails to confer long-term cell survival. Instead, it unveils robust alternative cell death pathways that function as backup mechanisms [86] [88]. This phenomenon has critical implications for drug development, suggesting that combinatorial strategies targeting multiple death pathways may be necessary for effective therapeutic intervention.
The core objective of this guide is to provide a comparative analysis of cell death pathways exposed by caspase inhibition, framing the discussion within the broader thesis that morphological features and biochemical pathways possess a high degree of specificity that can differentiate apoptosis from necrosis and other forms of cell death. We summarize key experimental data, provide detailed methodologies for pivotal experiments, and visualize the complex signaling crosstalk that governs cellular fate.
Comparative Morphology and Biochemistry of Cell Death Pathways
The table below provides a detailed comparison of the defining characteristics of different cell death modalities, with an emphasis on features that become prominent upon caspase inhibition.
Table 1: Comparative Analysis of Cell Death Pathways
| Feature | Apoptosis | Necrosis (Accidental) | Necroptosis (Regulated Necrosis) | Autophagic Cell Death |
|---|---|---|---|---|
| Regulation | Programmed, highly regulated [63] [89] | Accidental, unregulated [90] [89] | Programmed, regulated [34] | Programmed, can be regulated [86] |
| Morphology | Cell shrinkage, nuclear condensation (pyknosis), apoptotic bodies [63] [89] | Cell swelling (oncosis), organelle disruption, membrane rupture [90] [89] | Cell swelling, membrane rupture, but regulated initiation [34] | Accumulation of double-membrane autophagic vacuoles [86] |
| Membrane Integrity | Maintained until late stages (blebbing) [89] | Lost early [90] | Lost after MLKL activation [34] | Generally maintained until late stages |
| Inflammatory Response | Minimal ('silent'), no DAMP release [91] [34] | Potent, passive release of DAMPs [63] [34] | Potent, active release of DAMPs [34] | Variable, can be immunogenic |
| Caspase Dependence | Absolutely dependent (initiator & executioner caspases) [63] [89] | Caspase-independent [90] | Caspase-8 inhibition is a key trigger [91] [34] | Can be induced or sensitized by caspase inhibition (e.g., zVAD-fmk) [86] |
| Key Molecular Mediators | Caspase-3/8/9, Bcl-2 family, Apaf-1 [92] [89] | N/A (non-specific) | RIPK1, RIPK3, MLKL [91] [34] | RIP1, Beclin-1, LC3, negative role of caspase-8 [86] |
| Effect of Caspase Inhibition (e.g., zVAD-fmk) | Blocks apoptosis | Can sensitize to or potentiate necrosis [86] [93] | Unmasks and promotes this pathway [91] [34] | Can induce or sensitize to this pathway [86] |
Key Alternative Pathways Unmasked by Caspase Inhibition
Necroptosis
Experimental Evidence: In L929 fibrosarcoma cells, Tumor Necrosis Factor (TNF) typically induces necrosis. The addition of the caspase inhibitor zVAD-fmk was found to potentiate, rather than inhibit, this TNF-induced necrotic death [93]. This paradoxical sensitization led to the discovery of a regulated pathway, now termed necroptosis.
Molecular Mechanism: The key molecular switch involves the kinase RIP1. In the presence of caspase-8 activity, RIP1 is cleaved and inactivated, favoring apoptosis. When caspase-8 is inhibited (e.g., by zVAD-fmk), RIP1 is stabilized and interacts with RIP3 to form the necrosome. This complex phosphorylates the effector protein MLKL, which then oligomerizes and translocates to the plasma membrane, causing membrane permeabilization and the release of DAMPs [91] [34]. This pathway highlights the critical role of caspase-8 as a negative regulator of necrosis.
Autophagic Cell Death
Experimental Evidence: Studies using the broad-spectrum caspase inhibitor zVAD-fmk have demonstrated that it can induce a form of cell death characterized by the massive accumulation of autophagic vacuoles [86]. This is not a survival mechanism but a cytotoxic process.
Molecular Mechanism: The underlying mechanism involves RIP1, which also plays a role in this pathway. Furthermore, caspase inhibition appears to remove a suppressive signal on the autophagic process. Initially, autophagy may serve as a survival attempt by clearing damaged mitochondria and other components. However, when caspase activity is absent and the process occurs in excess, it becomes cytotoxic, leading to autophagic cell death [86].
The PARP-1 Switch Mechanism
Experimental Evidence: Research in L929 cells showed that CD95 ligation induced apoptosis, which was blocked by zVAD-fmk. In contrast, TNF-induced necrosis was potentiated by zVAD-fmk [93]. The key difference was found in the handling of the DNA repair enzyme PARP-1.
Molecular Mechanism: During successful apoptosis, executioner caspases (e.g., caspase-3) cleave and inactivate PARP-1, preserving cellular ATP for the energy-dependent apoptotic process. In TNF-induced signaling, if caspases are inhibited (e.g., by zVAD-fmk), PARP-1 becomes overactivated by cellular stresses like DNA damage. This leads to catastrophic depletion of NAD+ and ATP, shifting the cell's fate from apoptosis to energy-deficient necrosis [93]. This positions PARP-1 cleavage as a critical molecular switch between the two modes of death.
Detailed Experimental Protocols for Key Findings
Protocol: Demonstrating zVAD-fmk Sensitization to Necroptosis
This protocol is adapted from studies investigating TNF-mediated death in L929 cells [93].
1. Cell Culture and Treatment:
- Cell Line: Mouse L929 fibrosarcoma cells.
- Culture Conditions: Maintain in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum at 37°C in 5% CO₂.
- Experimental Groups:
- Group 1: Control (vehicle only)
- Group 2: TNFα (e.g., 10-100 ng/mL)
- Group 3: zVAD-fmk (e.g., 20-50 µM)
- Group 4: TNFα + zVAD-fmk
- Incubation Time: Treat cells for 6-24 hours before analysis.
2. Cell Viability Assessment:
- Method: Propidium Iodide (PI) uptake assay measured by flow cytometry.
- Principle: PI is a DNA dye that is excluded from live cells with intact membranes. Cells undergoing necroptosis lose membrane integrity and become PI-positive.
- Procedure: Harvest cells, resuspend in PBS containing PI (e.g., 1 µg/mL), and analyze immediately by flow cytometry. The percentage of PI-positive cells indicates non-viable, necrotic cells.
3. Key Expected Outcome:
- Group 4 (TNFα + zVAD-fmk) is expected to show a significantly higher percentage of PI-positive cells compared to Groups 2 and 3, demonstrating the sensitization to necrosis upon caspase inhibition [93].
Protocol: Assessing the Role of PARP-1
This protocol extends the previous experiment to investigate the metabolic switch [93].
1. Cell Treatment:
- Use the same cell line and treatment groups as in Protocol 4.1.
- Include an additional control group using a specific PARP inhibitor (e.g., 3-aminobenzamide, 3-AB at 1-5 mM) in combination with TNFα and zVAD-fmk.
2. ATP Measurement:
- Method: Use a commercial luciferase-based ATP assay kit.
- Procedure: Lyse cells at the end of the treatment period. Mix the cell lysate with the luciferase reagent, which emits light in proportion to the ATP concentration. Measure luminescence with a plate reader.
- Expected Outcome: Group 4 (TNFα + zVAD-fmk) will show a dramatic depletion of ATP compared to other groups. This ATP depletion and the associated necrosis will be rescued in the group co-treated with the PARP inhibitor [93].
3. PARP-1 Cleavage Analysis:
- Method: Western Blotting.
- Antibodies: Use an antibody that detects full-length PARP-1 (116 kDa) and its signature caspase-derived cleavage fragment (85 kDa).
- Expected Outcome: In Group 2 (TNFα alone), if some apoptosis occurs, the 85 kDa fragment may be visible. In Group 4 (TNFα + zVAD-fmk), cleavage will be absent, and only the full-length PARP-1 will be detected, correlating with its overactivation.
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Studying Alternative Cell Death Pathways
| Reagent / Tool | Function / Specificity | Key Experimental Use |
|---|---|---|
| zVAD-fmk (Pan-caspase inhibitor) | Irreversible, broad-spectrum caspase inhibitor [86] [87] | Foundational tool for unmasking alternative death pathways like necroptosis and autophagic death [86] [93] |
| Q-VD-OPh (Pan-caspase inhibitor) | Irreversible, broad-spectrum inhibitor; less toxic than zVAD-fmk in vivo [87] | Used for long-term in vivo studies to inhibit caspases with reduced off-target effects [87] |
| Necrostatin-1 (Nec-1) | Specific allosteric inhibitor of RIPK1 kinase activity [34] | Used to confirm the role of RIPK1 and necroptosis in a death process; can distinguish from other pathways. |
| Propidium Iodide (PI) | Fluorescent DNA dye impermeable to live cells [93] | Standard flow cytometry assay to quantify necrotic/late apoptotic cells with compromised membranes [93] |
| Anti-phospho-MLKL Antibody | Detects the activated (phosphorylated) form of MLKL [34] | Gold-standard immunoblot or immunofluorescence method to specifically confirm ongoing necroptosis. |
| LC3 Antibody | Detects lipidated form of LC3 (LC3-II) associated with autophagosomes | Used in Western blot or immunofluorescence to monitor autophagy induction. |
| PARP-1 Antibody | Detects both full-length (116 kDa) and cleaved (85 kDa) PARP-1 [93] | Key readout for caspase activity and the molecular switch between apoptosis and necrosis [93] |
| Recombinant TNFα | Potent activator of TNFR1, can induce both apoptosis and necroptosis [93] [34] | Standard ligand used in combination with caspase inhibitors to trigger and study necroptosis in vitro. |
The experimental use of caspase inhibitors has been instrumental in deconstructing the simplistic apoptosis-necrosis dichotomy, revealing a complex network of interconnected cell death pathways with distinct morphological and biochemical signatures. The data clearly show that caspase inhibition can act as a double-edged sword: while it effectively blocks apoptosis, it simultaneously removes critical inhibitory brakes on alternative pathways like necroptosis.
For researchers and drug development professionals, these findings carry profound implications. Therapeutic strategies aimed at modulating cell death must move beyond targeting a single pathway. For instance, in acute lung injury, where PANoptosis (a combined form of apoptosis, necroptosis, and pyroptosis) occurs, inhibiting a single pathway like apoptosis may be insufficient [91]. Instead, combinational approaches targeting core regulatory nodes, such as caspase-8 along with RIPK1 or RIPK3, may yield greater efficacy. The continued development of specific inhibitors for necroptosis (e.g., Nec-1) and other alternative pathways, coupled with a deeper understanding of their crosstalk, will be crucial for designing the next generation of cytoprotective therapies.
Impact of Cell Culture Conditions and Sample Handling on Morphology
In the field of cell biology and drug development, accurately distinguishing between apoptosis and necrosis is paramount, as the mechanism of cell death has significant implications for understanding compound toxicity, therapeutic efficacy, and disease pathology. The morphological features of dying cells serve as a primary endpoint for this discrimination. However, these morphological specifics are not determined solely by the death stimulus; they are profoundly influenced by cell culture conditions and sample handling protocols. Environmental factors such as nutrient availability, metabolic by-product accumulation, and physical handling can induce stress, altering the canonical death pathways and leading to misinterpretation. This guide objectively compares the morphological outcomes under varying culture and handling conditions, providing a structured framework to enhance the reliability of cell death research.
The table below summarizes the core morphological characteristics that distinguish apoptosis from necrosis, which form the basis for visual analysis in research.
Table 1: Key Morphological Differences Between Apoptosis and Necrosis
| Feature | Apoptosis | Necrosis |
|---|---|---|
| Overall Cell Shape | Cell shrinkage, rounding, and loss of cell-cell contacts [94] [95]. | Cell swelling (oncosis) leading to rupture [94] [95]. |
| Plasma Membrane | Blebbing with intact integrity; formation of membrane-bound apoptotic bodies [94] [95]. | Loss of integrity and increased permeability; rapid membrane rupture [94] [7] [95]. |
| Nucleus | Chromatin condensation (pyknosis) and fragmentation into oligonucleosomal pieces (DNA laddering) [63] [94]. | Random degradation of genomic DNA; condensation and disintegration [94] [95]. |
| Cellular Organelles | Generally no visible changes to organelles in early stages [95]. | Swelling of mitochondria and endoplasmic reticulum; decay of the Golgi apparatus [94] [95]. |
| Post-Death Clearance | Phagocytosis by neighboring macrophages [63] [95]. | Cell lysis; release of intracellular contents [94] [95]. |
| Inflammatory Response | Typically none (immunologically silent) [63] [96]. | Significant inflammatory response triggered [96] [7] [95]. |
| Energy Dependence | ATP-dependent process [95]. | ATP-independent process [95]. |
The Impact of Culture Conditions on Cell Death Morphology
Cell culture environment is a critical determinant of cell health and death pathway. Deviations from optimal conditions can stress cells, pushing them toward unintended death modalities.
- Nutrient Depletion and By-product Accumulation: In high-density cultures, the depletion of essential nutrients and the accumulation of metabolic by-products like lactate and ammonia constitute a significant environmental perturbation [94]. This stress can upregulate pro-apoptotic proteins, leading to caspase activation and apoptosis. However, severe and sustained stress can overwhelm the regulated apoptotic program, resulting in a shift to necrosis [94].
- Serum Concentration and Growth Factors: Serum deprivation is a well-known activator of the intrinsic apoptotic pathway, leading to characteristic cell shrinkage and DNA fragmentation [97]. The withdrawal of survival signals triggers mitochondrial outer membrane permeabilization (MOMP), a key step in apoptosis [96].
- Oxygen Levels (Hypoxia): Perturbations in oxygen transport, common in dense cultures or inadequate bioreactor systems, can induce endoplasmic reticulum (ER) stress. This can activate the intrinsic ER stress pathway, leading to apoptosis via caspase-12, or under prolonged stress, trigger necrotic cell death [94].
Sample Handling and Its Effects on Morphological Integrity
The methods used to manipulate and analyze cells can introduce artifacts that obscure true morphological signatures.
- Detachment Techniques: Overly aggressive trypsinization or mechanical scraping can damage the plasma membrane. This can cause immediate necrotic-like rupture or induce secondary apoptosis in culture post-handling.
- Fixation Timing and Quality: Delayed or improper fixation can allow degradative processes to continue ex vivo, blurring the distinct morphological features. For instance, late-stage apoptotic cells can secondaryly lose membrane integrity, resembling necrosis [96].
- Imaging and Analysis: The choice of imaging technique is crucial. Label-free, non-invasive methods like Full-Field Optical Coherence Tomography (FF-OCT) allow for the real-time monitoring of dynamic processes like membrane blebbing in apoptosis or rapid swelling in necrosis without introducing staining artifacts [7]. In contrast, techniques that require fixation and staining preclude real-time analysis and can sometimes affect structural integrity [7].
Experimental Protocols for Inducing and Analyzing Cell Death
To generate reliable and comparable data, standardized protocols for inducing death and analyzing morphology are essential.
Protocol 1: Inducing and Imaging Apoptosis with Doxorubicin
This protocol uses a chemotherapeutic agent to trigger intrinsic apoptosis and FF-OCT for label-free observation [7].
- Cell Preparation: Culture HeLa cells (or relevant cell line) as a monolayer in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with serum under standard conditions (37°C, 5% CO₂).
- Apoptosis Induction: Add doxorubicin to the culture medium at a final concentration of 5 μmol/L. Doxorubicin intercalates into DNA, causing double-strand breaks and activating the p53 pathway [7].
- Real-Time Imaging: Immediately transfer the culture dish to a time-domain FF-OCT system. This system uses a broadband halogen light source and identical 40x water-immersion objectives in a Linnik interferometer configuration to achieve sub-micrometer resolution.
- Data Acquisition: Acquire images continuously at 20-minute intervals for up to 180 minutes. The system generates en face (x-y) cross-sectional images by processing phase-shifted interference images, which are then stacked to reconstruct 3D cellular topography [7].
- Morphological Analysis: Identify key apoptotic features: echinoid spine formation, cell contraction, membrane blebbing, and filopodia reorganization [7].
Protocol 2: Inducing and Imaging Necrosis with Ethanol
This protocol uses a high-concentration chemical insult to induce rapid, unplanned necrosis [7].
- Cell Preparation: Culture HeLa cells as described in Protocol 1.
- Necrosis Induction: Treat cells with 99% ethanol. Ethanol's lipophilic nature disrupts the phospholipid bilayer, denatures proteins, and impairs ion homeostasis, leading to rapid oncosis [7].
- Real-Time Imaging: Use the same FF-OCT system and imaging parameters as in Protocol 1.
- Morphological Analysis: Identify key necrotic features: rapid membrane rupture, intracellular content leakage, and abrupt loss of adhesion structure [7].
Research Reagent Solutions for Cell Death Studies
The table below lists essential reagents and their functions for studying cell death morphology.
Table 2: Key Research Reagents and Their Applications
| Reagent | Function in Research |
|---|---|
| Doxorubicin | A chemotherapeutic agent used to induce the intrinsic pathway of apoptosis by causing DNA damage [7]. |
| Ethanol | Used at high concentrations (e.g., 99%) to induce necrosis by disrupting membrane integrity and denaturing proteins [7]. |
| Caspase-3 Antibody | An immunohistochemistry tool to detect activated caspase-3, a key executioner protease, serving as a biochemical marker for apoptosis [95]. |
| BAX Antibody | Used to detect the conformational change and mitochondrial translocation of BAX, a pro-apoptotic Bcl-2 protein, during intrinsic apoptosis [95]. |
| RIP3 & MLKL Antibodies | Essential reagents for detecting key mediators of necroptosis, a regulated form of necrotic cell death, via techniques like immunoprecipitation and immunofluorescence [95]. |
| Serum-Free Media | Used to study death receptor-mediated (extrinsic) apoptosis or to create nutrient stress that can induce autophagy or apoptosis [94]. |
| Annexin V | Binds to phosphatidylserine (PS) exposed on the outer leaflet of the plasma membrane, an early event in apoptosis used in flow cytometry [63]. |
Signaling Pathways in Apoptosis and Necroptosis
The following diagrams, generated using Graphviz DOT language, illustrate the core signaling pathways governing apoptosis and necroptosis, highlighting key molecular players that can be influenced by cellular stress.
The accurate interpretation of cell death morphology is a cornerstone of biomedical research, but it is a process highly susceptible to external influences. Cell culture conditions—ranging from nutrient levels and metabolic waste to oxygenation—and sample handling procedures—from detachment to fixation—can significantly alter the morphological presentation of apoptosis and necrosis. By adhering to standardized, carefully controlled experimental protocols and utilizing advanced, label-free imaging techniques, researchers can minimize artifacts and obtain more reliable data. A rigorous understanding of these factors ensures that morphological assessment remains a specific and powerful tool for differentiating cell death mechanisms in drug development and basic biological research.
Optimizing Protocols to Minimize Artifacts in Imaging and Analysis
Accurately distinguishing between apoptosis and necrosis is fundamental to biomedical research, influencing areas from anticancer therapy development to neurodegenerative disease studies. A core challenge in this field lies in the accurate capture and interpretation of cellular morphology, a process often compromised by technical artifacts inherent to the imaging and analytical techniques themselves. This guide provides a objective comparison of contemporary methods for visualizing cell death, focusing on their specific vulnerabilities to artifacts and the optimized protocols that can mitigate them. By comparing the performance of label-free imaging, fluorescent reporter systems, and flow cytometry, we aim to provide researchers with the data necessary to select the most appropriate method, thereby enhancing the specificity and reliability of morphological data in apoptosis versus necrosis research.
Comparative Analysis of Cell Death Imaging Modalities
The choice of imaging technology significantly impacts the type and severity of artifacts encountered. The table below compares the performance of four key technologies used in cell death research.
Table 1: Performance Comparison of Cell Death Imaging and Analysis Modalities
| Technology | Key Principle | Spatial Resolution | Key Artifacts & Limitations | Optimal Use Case |
|---|---|---|---|---|
| FF-OCT [7] | Label-free interferometry with broadband light source | <1 µm axial and transverse [7] | - Limited molecular specificity without labels.- Requires specialized custom-built systems. | High-resolution 3D morphological analysis of apoptosis vs. necrosis. |
| Fluorescence Microscopy (FM) [98] | Widefield detection of fluorophore emission | ~200 nm (diffraction-limited) [98] | - Photobleaching & phototoxicity [98].- Sample autofluorescence & background scatter [98].- Shallow depth of field [98]. | Direct visualization of cell morphology and viability in adherent cultures. |
| Flow Cytometry (FCM) [98] | Laser-based scattering and fluorescence of suspended cells | N/A (single-cell analysis) | - Requires cell detachment, risking selective loss [98].- Cannot visualize spatial relationships. | High-throughput, quantitative viability and death mechanism subtyping. |
| Caspase-3/-7 Reporter [99] | Caspase-dependent reconstitution of split-GFP | Limited by host microscope | - Reporter expression variability.- Potential delay between activation and signal. | Real-time, dynamic tracking of apoptotic executioner activity in live cells. |
Detailed Methodologies for Key Imaging Protocols
Protocol: Full-Field Optical Coherence Tomography (FF-OCT) for Label-Free Morphology
FF-OCT is a label-free, non-invasive imaging technique that enables high-resolution visualization of cellular structural changes, making it ideal for distinguishing apoptosis and necrosis based on morphology [7].
- Cell Preparation and Death Induction: HeLa cells are cultured as a monolayer in DMEM under standard conditions (5% CO₂, 37°C) [7].
- Apoptosis Induction: Treat cells with 5 μmol/L doxorubicin in 1.5 mL culture medium. Doxorubicin intercalates into DNA and inhibits topoisomerase II, causing double-strand breaks and activating p53-mediated apoptosis [7].
- Necrosis Induction: Treat cells with 99% ethanol. Ethanol's lipophilic nature disrupts the phospholipid bilayer, denatures proteins, and causes rapid loss of membrane integrity, leading to necrosis [7].
- Imaging: Initiate FF-OCT imaging immediately post-treatment and continue at 20-minute intervals for up to 180 minutes [7].
- FF-OCT System Configuration: A custom-built time-domain FF-OCT system is used [7].
- Light Source: Broadband halogen lamp (center wavelength: 650 nm, spectral width: 200 nm) for sub-micrometer axial resolution [7].
- Interferometer: Linnik-configured Michelson interferometer with identical 40x water-immersion objectives (NA: 0.8) in reference and sample arms for subcellular resolution [7].
- Detection: A CCD camera (1024 x 1024 pixels, 12-bit, 20 fps) captures 2D interference images. Phase shifting is achieved via a piezoelectric actuator on the reference mirror [7].
- Image Processing and 3D Reconstruction: Temporally phase-shifted images are processed to remove the DC component, isolating sample reflection data. A stack of these en face (x-y) cross-sectional images is compiled using a motorized z-stage to reconstruct 3D volume and surface topography [7].
Protocol: Fluorescent Reporter System for Caspase-3/-7 Dynamics
This protocol uses a stable fluorescent reporter system for real-time imaging of executioner caspase activity, a key marker of apoptosis [99].
- Reporter Cell Line Generation:
- Construct Design: A lentiviral vector is used to deliver a cassette encoding a ZipGFP-based caspase-3/-7 reporter and a constitutive mCherry marker. The ZipGFP reporter contains a caspase-specific DEVD cleavage motif within a split-GFP architecture [99].
- Mechanism: In healthy cells, the split-GFP fragments are held together, preventing fluorescence. During apoptosis, caspase-3/-7 cleaves the DEVD linker, allowing GFP to refold and emit a persistent fluorescent signal. The mCherry signal serves as a cell presence control [99].
- Live-Cell Imaging of Apoptosis:
- Induction: Treat stable reporter cells with an apoptosis inducer (e.g., 1-10 µM carfilzomib or 50-100 µM oxaliplatin). To confirm caspase-dependency, include a control with a pan-caspase inhibitor (e.g., 20-50 µM zVAD-FMK) [99].
- Image Acquisition: Perform time-lapse imaging over 48-120 hours using a fluorescence microscope equipped with environmental control (37°C, 5% CO₂). Acquire GFP and mCherry channels at regular intervals (e.g., every 30-60 minutes) [99].
- Validation: Validate apoptosis induction in parallel samples via western blotting for cleaved PARP and cleaved caspase-3, or by flow cytometry with Annexin V/PI staining [99].
- Application in 3D Models: The reporter system can be adapted to 3D spheroids or patient-derived organoids (PDOs). Seed reporter cells in Cultrex or other ECM substitutes to form 3D structures. Treat and image as above, ensuring adequate z-stack acquisition to capture the 3D volume [99].
Protocol: Comparative Viability Assessment via FM and FCM
This protocol directly compares fluorescence microscopy and flow cytometry for assessing cytotoxicity, highlighting methodological strengths and artifacts [98].
- Cell Culture and Cytotoxic Treatment:
- Cells and Material: Use SAOS-2 osteoblast-like cells. The cytotoxic agent is Bioglass 45S5 (BG) particles, sieved into three size ranges (<38 µm, 63–125 µm, and 315–500 µm) at concentrations of 25, 50, and 100 mg/mL [98].
- Treatment: Treat cells with BG particles for 3 and 72 hours. Include an untreated control. Monitor pH changes in the medium, as BG dissolution increases pH, contributing to cytotoxic stress [98].
- Fluorescence Microscopy (FM) Protocol:
- Staining: Stain cells with a FDA/PI (fluorescein diacetate/propidium iodide) mixture. FDA is metabolized by live cells to produce green fluorescence, while PI is a red fluorescent DNA dye excluded by live cells but entering dead cells [98].
- Image Acquisition and Analysis: Capture multiple random fields per sample using a widefield fluorescence microscope. Manually or automatically count viable (FDA+) and nonviable (PI+) cells. Viability is calculated as (FDA+ cells / total cells) × 100 [98].
- Flow Cytometry (FCM) Protocol:
- Staining and Cell Harvest: Use a multiparametric staining kit (e.g., containing Hoechst, DiIC1, Annexin V-FITC, and PI). Harvest cells into suspension, ensuring a single-cell suspension [98].
- Data Acquisition and Analysis: Acquire data on a flow cytometer, analyzing a minimum of 10,000 events per sample. Use FSC/SSC to gate on single cells. Classify populations as:
- Viable: Hoechst+, Annexin V-, PI-.
- Early Apoptotic: Hoechst+, Annexin V+, PI-.
- Late Apoptotic/Necrotic: Hoechst+, Annexin V+, PI+ [98].
- Data Correlation: Plot viability percentages obtained from FM against those from FCM to assess correlation (e.g., Pearson correlation coefficient r). Expect a strong correlation (r > 0.9), though FCM typically shows greater precision, especially at low viability [98].
Visualizing Signaling Pathways and Experimental Workflows
Apoptotic Signaling and Caspase-3 Activation Pathway
The diagram below illustrates the key pathways leading to caspase-3 activation, a central event in apoptosis that can be tracked with specific reporters [99] [100].
Comparative Experimental Workflow: FM vs. FCM
This workflow diagram outlines the parallel processes for preparing and analyzing samples using fluorescence microscopy and flow cytometry, highlighting key steps where artifacts can be introduced [98].
Research Reagent Solutions for Cell Death Analysis
The table below catalogs key reagents and their functions for studying apoptosis and necrosis, helping researchers select appropriate tools for their experimental protocols.
Table 2: Essential Research Reagents for Apoptosis and Necrosis Analysis
| Reagent / Tool | Function / Target | Key Characteristics & Applications |
|---|---|---|
| Doxorubicin [7] | Apoptosis inducer (chemical) | DNA intercalator; inhibits topoisomerase II; activates p53 pathway. Used at ~5 µM to induce apoptosis in HeLa cells [7]. |
| Ethanol (99%) [7] | Necrosis inducer (chemical) | Disrupts phospholipid bilayer and denatures proteins; induces rapid, uncontrolled necrosis at high concentrations [7]. |
| ZipGFP Caspase-3/-7 Reporter [99] | Fluorescent biosensor for executioner caspases | Split-GFP with DEVD cleavage motif; low background, irreversible fluorescence upon caspase activation. For real-time apoptosis tracking in 2D/3D models [99]. |
| Annexin V / Propidium Iodide (PI) [99] [98] | Flow cytometry / microscopy stains | Binds phosphatidylserine (externalized in apoptosis); PI stains DNA in dead cells with compromised membranes. Distinguishes live, early apoptotic, and late apoptotic/necrotic cells [99] [98]. |
| FDA / PI [98] | Fluorescence microscopy viability stain | FDA metabolized to green fluorescein in live cells; PI enters dead cells (red). Standard for live/dead assessment via FM [98]. |
| zVAD-FMK [99] | Pan-caspase inhibitor | Irreversible inhibitor; used as a control (e.g., 20-50 µM) to confirm caspase-dependent processes in reporter assays [99]. |
| Isatin Sulfonamide Probes [100] | Radiotracer / activity-based probe for caspase-3/7 | Small molecule (e.g., ICMT-11) with high affinity (IC50 ~0.5 nM); forms reversible covalent bond. Basis for caspase-3 PET radiotracers [100]. |
A fundamental challenge in oncology is the heterogeneous response of cancers to therapeutic agents. This case study delves into the critical task of analyzing drug-induced cell death, focusing on the distinct morphological and biochemical pathways of apoptosis and necrosis. Accurately differentiating between these mechanisms is not merely academic; it provides vital insights into a drug's mechanism of action (MOA), its efficacy, and potential side effects, thereby guiding subsequent research and development choices [101] [102]. We present a comparative analysis of cell death pathways activated by different stimuli across various cancer cell lines, providing a structured guide with quantitative data, experimental protocols, and key reagents to support research in this field.
Comparative Analysis of Cell Death Pathways
Key Features of Apoptosis, Necrosis, and Related Pathways
Table 1: Characteristics of Major Cell Death Pathways
| Feature | Apoptosis | Necroptosis | Pyroptosis | Ferroptosis |
|---|---|---|---|---|
| Morphology | Cell shrinkage, chromatin condensation, blebbing | Cell swelling, plasma membrane rupture | Cell swelling, pore formation, membrane lysis | Shrunken mitochondria, loss of cristae |
| Key Regulators | Caspases-3/8/9, Bcl-2/Bax [44] | RIPK1, RIPK3, MLKL [102] | Gasdermin family, Caspase-1 [102] | GPX4, Lipid ROS [103] |
| Biomarkers | Annexin V/PI, cleaved Caspase-3 [104] | p-MLKL [102] | GSDMD-N, IL-1β [102] | Lipid peroxides (C11-BODIPY) [103] |
| Inflammatory Response | Non-inflammatory | Inflammatory | Highly inflammatory | Immunogenic |
Quantitative Comparison of Drug Responses in Lung Cancer Cell Lines
A study investigating hematoporphyrin derivative-mediated photodynamic therapy (HPD-PDT) on human lung cancer cell lines revealed significant variations in cell death responses, underscoring the importance of cell line-specific profiling [101].
Table 2: HPD-PDT Response in Lung Cell Lines (Adapted from Ma et al.)
| Cell Line | Cell Type | Viability Trend | Relative ROS Production | Relative Apoptosis Rate | Key Protein Changes |
|---|---|---|---|---|---|
| H520 | Lung squamous carcinoma | Gradual decrease with HPD dose | Highest | Highest | Reduced Bcl-2, upregulated Bax, activated Caspase-3 |
| A549 | Lung adenocarcinoma | Gradual decrease with HPD dose | High | High | Reduced Bcl-2, upregulated Bax, activated Caspase-3 |
| H446 | Lung small cell carcinoma | Gradual decrease with HPD dose | Moderate | Moderate | Reduced Bcl-2, upregulated Bax, activated Caspase-3 |
| BEAS-2B | Normal bronchial epithelial | Gradual decrease with HPD dose | Lowest | Lowest | Reduced Bcl-2, upregulated Bax, activated Caspase-3 |
The study confirmed that HPD-PDT induced apoptosis and autophagy across all lines, but the extent varied, with cancer cells generally showing higher susceptibility than normal cells. This highlights that lung cancer cells die via interactions of different death pathways rather than a single, uniform mechanism [101].
Experimental Protocols for Cell Death Analysis
Protocol 1: Assessing Apoptosis via Multiparametric Flow Cytometry
This protocol is adapted from studies on peripheral blood mononuclear cells (PBMCs) and is widely applicable for quantifying apoptosis in cell lines [104].
- 1. Sample Preparation: Harvest and wash cells. For suspension cells, proceed directly. For adherent cells, use gentle enzymatic (e.g., trypsin-EDTA) or non-enzymatic dissociation to preserve membrane integrity.
- 2. Staining: Resuspend 1 × 10^6 cells in a binding buffer.
- Annexin V/PI Staining: Add Annexin V-FITC and Propidium Iodide (PI) following kit instructions (e.g., BioLegend, Cat# 640914). Incubate for 15 minutes in the dark at room temperature [104].
- Intracellular Staining (if required): Fix and permeabilize cells using a commercial kit (e.g., Cytofix/Cytoperm). Stain with antibodies against pro-apoptotic proteins (e.g., Bax, Clone 2D2) and anti-apoptotic proteins (e.g., Bcl-2, Clone 100) [104].
- 3. Flow Cytometry Acquisition: Analyze samples on a flow cytometer (e.g., BD FACSCanto II), acquiring at least 10,000 events per sample. Use unstained and single-stained controls for compensation [104].
- 4. Data Analysis: Use software like FlowJo.
- Identify viable (Annexin V⁻/PI⁻), early apoptotic (Annexin V⁺/PI⁻), late apoptotic (Annexin V⁺/PI⁺), and necrotic (Annexin V⁻/PI⁺) populations.
- Calculate median fluorescence intensity (MFI) for intracellular markers and Bax/Bcl-2 ratios [104].
Protocol 2: Inducing and Validating PANoptosis in an Inflammatory Model
This protocol is based on research into TNF-α-induced cell death in MC3T3-E1 osteoblasts, a model applicable to studying complex inflammatory cell death in other lineages [102].
- 1. Cell Culture and Stimulation: Culture cells (e.g., MC3T3-E1) in appropriate medium (e.g., MEM-α with 10% FBS). To induce PANoptosis, stimulate cells with 10-50 ng/mL of TNF-α for 72 hours [102].
- 2. Inhibitor Treatment (Optional): To dissect pathways, pre-treat cells with specific inhibitors 30 minutes prior to TNF-α stimulation. For example, use 10 μM CY-09 to inhibit NLRP3 and suppress pyroptosis [102].
- 3. Cell Death Assessment:
- LDH Release Assay: Quantify lactate dehydrogenase (LDH) in the supernatant as a marker of plasma membrane damage.
- PI Staining: Stain cells with PI and visualize via fluorescence microscopy to identify dead cells with compromised membranes.
- Western Blotting: Confirm the activation of multiple death pathways by probing for:
- Apoptosis: Cleaved Caspase-3, Cleaved Caspase-8
- Necroptosis: p-RIPK1, p-MLKL
- Pyroptosis: Cleaved GSDMD [102]
- 4. Morphological Analysis: Use scanning electron microscopy (SEM) to identify mixed morphological features (e.g., apoptotic bodies alongside necrotic membrane rupture) in the same field of view, a hallmark of PANoptosis [102].
Signaling Pathways in Drug-Induced Cell Death
Apoptosis Signaling Pathway
Ferroptosis Initiation at Organelle Contact Sites
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Cell Death Analysis
| Reagent / Assay | Function / Target | Example Application |
|---|---|---|
| Annexin V / PI Apoptosis Kit | Binds phosphatidylserine (PS) on apoptotic cells; PI stains DNA in membrane-compromised cells. | Differentiating early/late apoptosis and necrosis via flow cytometry [104]. |
| C11-BODIPY 581/591 | Ratiometric fluorescent sensor for lipid peroxidation. Oxidized form shifts fluorescence. | Detecting lipid ROS during ferroptosis [103]. |
| Anti-Cleaved Caspase-3 Antibody | Detects activated Caspase-3, a key executioner protease. | Confirming engagement of apoptotic pathway via Western blot or flow cytometry [104] [102]. |
| JC-1 Dye | Mitochondrial membrane potential (ΔΨm) sensor. Emits red in high ΔΨm, green in low ΔΨm. | Measuring mitochondrial depolarization, an early event in intrinsic apoptosis [104]. |
| Anti-Bax & Anti-Bcl-2 Antibodies | Detect pro-apoptotic (Bax) and anti-apoptotic (Bcl-2) proteins. | Assessing Bax/Bcl-2 ratio, a key indicator of apoptotic susceptibility [101] [104]. |
| CY-09 | Specific inhibitor of the NLRP3 inflammasome. | Inhibiting pyroptosis to dissect its role in PANoptosis models [102]. |
| Ferrostatin-1 (Fer-1) | Potent radical-trapping antioxidant. | Specifically inhibiting ferroptosis by halting phospholipid peroxidation [103]. |
| Z-VAD-FMK | Pan-caspase inhibitor. | Inhibiting apoptosis to dissect its contribution in mixed cell death models [103]. |
Discussion and Research Implications
This case study underscores the complexity of drug-induced cell death, demonstrating that therapeutic responses are rarely confined to a single, pure pathway. The data reveals a spectrum of death modalities, from the classical apoptosis induced by targeted agents like Venetoclax [105] to the mixed PANoptosis phenotype triggered by inflammatory stimuli like TNF-α [102]. A critical finding from recent research is the role of organelle contact sites, such as ER-mitochondria connections, as key hubs for initiating lipid peroxidation and driving ferroptosis [103]. This spatial regulation of cell death adds another layer of sophistication to our understanding.
For researchers, the implication is clear: a multi-faceted analytical approach is essential. Relying on a single assay can lead to misinterpretation. Instead, combining flow cytometry for quantitation, Western blotting for pathway mapping, and advanced imaging for morphological context provides a holistic view. Furthermore, the development of machine learning models that integrate transcriptomic data from cell lines, such as the CellHit pipeline, shows great promise in predicting drug sensitivity and deconstructing the biological processes associated with a drug's mechanism of action [105]. As the field advances, incorporating these multi-omics and computational approaches with traditional cell death assays will be crucial for accelerating the development of more effective and precise cancer therapies.
Confirming the Pathway: Validation and Comparative Analysis of Cell Death Modalities
Cell death is a fundamental physiological process in all living organisms, with roles extending from embryonic development and organ maintenance to the coordination of immune responses and disease pathology [34]. For researchers, scientists, and drug development professionals, accurately distinguishing between different cell death modalities is crucial for understanding disease mechanisms and developing targeted therapies. Historically, cell death was simplistically dichotomized into apoptosis (programmed) and necrosis (accidental); however, advancements over recent decades have revealed a more complex landscape, including regulated forms of necrosis such as necroptosis [3] [34].
This guide provides a detailed, objective comparison of three key cell death pathways—apoptosis, necrosis, and necroptosis—focusing on their distinct morphological features, biochemical mechanisms, and functional consequences. The specificity of morphological characteristics is particularly vital for research, as it enables initial identification and differentiation of cell death types in experimental settings, guiding subsequent molecular analysis. We present summarized quantitative data, detailed experimental protocols for observation, and essential research tools to support rigorous investigation in this field.
The table below provides a side-by-side comparison of the defining features of apoptosis, necrosis, and necroptosis, synthesizing key morphological, biochemical, and functional data for easy reference.
Table 1: Comprehensive Comparison of Apoptosis, Necrosis, and Necroptosis
| Feature | Apoptosis | Necrosis | Necroptosis |
|---|---|---|---|
| Classification | Programmed Cell Death (PCD), Regulated Cell Death (RCD) [3] [106] | Accidental Cell Death (ACD) [3] [106] | Programmed Cell Death (PCD), Regulated Cell Death (RCD) [107] [108] |
| Primary Stimuli | Physiological signals during development, DNA damage, ER stress, growth factor deprivation [109] [110] | Extreme physical/chemical trauma, ischemia, infection, toxins [106] [107] [111] | Death receptor ligands (e.g., TNF-α), TLR ligands, Caspase-8 inhibition [108] [34] |
| Key Morphological Features | Cell shrinkage, chromatin condensation, nuclear fragmentation, membrane blebbing, formation of apoptotic bodies [3] [109] [106] | Cell and organelle swelling, loss of membrane integrity, plasma membrane rupture [3] [106] [107] | Cell swelling, plasma membrane rupture (similar to necrosis) but regulated [3] [108] [34] |
| Membrane Integrity | Maintained until late stages; membrane blebbing with intact integrity [106] [111] | Lost; increased permeability leading to rupture [106] [107] | Lost; MLKL pores disrupt membrane, leading to rupture [108] [34] |
| Inflammatory Response | Typically non-inflammatory ("immunologically silent") [109] [34] | Strongly inflammatory due to release of intracellular contents [106] [34] | Highly immunogenic and inflammatory [107] [34] |
| Key Molecular Mediators | Caspases (e.g., Caspase-3/8/9), BCL-2 family, APAF-1, Cytochrome c [3] [109] [110] | Non-specific; often involves ATP depletion, calcium influx, and lysosomal hydrolases [106] [107] | RIPK1, RIPK3, MLKL [108] [34] |
| Caspase Dependence | Caspase-dependent [3] [110] | Caspase-independent [106] [107] | Caspase-independent [107] [34] |
| Energy Dependency | ATP-dependent [106] [111] | ATP-independent [106] [111] | Presumed ATP-dependent (regulated process) |
| Post-Death Clearance | Phagocytosis by neighboring cells [109] [106] | Cell lysis; phagocytosis by immune cells after inflammatory response [106] [107] | Cell lysis; release of DAMPs triggers immune response [107] [34] |
Morphological and Molecular Mechanisms
Characteristic Morphological Changes
The definitive identification of cell death types relies heavily on observing specific morphological characteristics, which are best visualized using high-resolution imaging techniques.
- Apoptosis: The cell undergoes a controlled, step-wise dismantling. Key morphological hallmarks include cell shrinkage and loss of cell-cell contacts, chromatin condensation (pyknosis) and nuclear fragmentation (karyorrhexis), blebbing of the plasma membrane with the formation of small, membrane-bound apoptotic bodies, and phagocytosis of these bodies by neighboring macrophages or parenchymal cells without eliciting an inflammatory response [3] [109] [106]. The integrity of the plasma membrane is maintained until the final stages, preventing the release of pro-inflammatory intracellular components [111].
- Necrosis: This is a chaotic and passive process characterized by a loss of regulatory control. Morphological signs include rapid cell and organelle swelling (oncosis), disruption of the plasma membrane and loss of integrity, swelling and rupture of mitochondria and the endoplasmic reticulum, and eventual cell lysis. The release of cellular contents into the extracellular space acts as a potent trigger for a strong inflammatory response [3] [106] [107].
- Necroptosis: While morphologically resembling necrosis with features like cell swelling and plasma membrane rupture, necroptosis is molecularly regulated. The rupture is executed by the oligomerization of phosphorylated MLKL, which forms pores in the plasma membrane [108] [34]. Like necrosis, the lytic nature of necroptosis results in the release of damage-associated molecular patterns (DAMPs) and a consequent immunogenic response [107] [34].
Underlying Signaling Pathways
The distinct morphological outcomes are determined by dedicated and well-characterized molecular pathways.
Apoptosis Pathways: Apoptosis proceeds via two main pathways that converge on caspase activation.
- The Extrinsic Pathway is initiated by the binding of extracellular death ligands (e.g., FasL, TNF-α) to cell surface death receptors. This leads to the formation of the Death-Inducing Signaling Complex (DISC), which recruits and activates initiator caspase-8. Active caspase-8 can directly cleave and activate executioner caspases like caspase-3/7 [109] [110].
- The Intrinsic Pathway is triggered by internal cellular stresses (e.g., DNA damage, oxidative stress). These signals cause the BCL-2 family proteins BAX and BAK to permeabilize the mitochondrial outer membrane (MOMP). This leads to the release of cytochrome c, which forms the apoptosome with APAF-1 and activates caspase-9, which in turn activates executioner caspases [3] [109] [110].
The following diagram illustrates the key steps in these apoptotic pathways:
Diagram 1: Key signaling pathways in extrinsic and intrinsic apoptosis.
Necroptosis Pathway: Necroptosis serves as a backup cell death pathway when apoptosis is blocked, particularly when caspase-8 is inhibited. It can be initiated by death receptors like TNFR1. Upon ligand binding and under conditions of caspase-8 inhibition, RIPK1 and RIPK3 interact via their RHIM domains, forming a complex called the necrosome. RIPK3 then phosphorylates the effector protein MLKL. Phosphorylated MLKL oligomerizes, translocates to the plasma membrane, and forms pores, leading to membrane rupture and the release of inflammatory cellular contents [108] [34].
The molecular cascade of necroptosis is detailed below:
Diagram 2: The molecular pathway of TNF-α-induced necroptosis.
Experimental Protocols for Cell Death Observation
High-Resolution Imaging of Morphological Changes
A 2025 study demonstrated the use of label-free, high-resolution imaging to distinguish apoptosis and necrosis in real-time [7]. The protocol below is adapted from this research.
- Objective: To monitor and distinguish morphological alterations in living cells undergoing apoptosis or necrosis without the use of labels.
- Cell Line and Culture: HeLa cells (human cervical cancer cells) are cultured as a monolayer in Dulbecco’s Modified Eagle’s Medium (DMEM) under standard conditions (37°C, 5% CO₂) [7].
- Induction of Cell Death:
- Apoptosis Induction: Treat cells with 5 μmol/L Doxorubicin. Doxorubicin is an anthracycline chemotherapeutic that intercalates into DNA and inhibits topoisomerase II, causing double-strand breaks and activating the p53 pathway, leading to apoptosis [7].
- Necrosis Induction: Treat cells with a high concentration (e.g., 99%) of Ethanol. Ethanol acts as a lipophilic agent that rapidly disrupts the phospholipid bilayer, denatures proteins, impairs ion homeostasis, and causes cell swelling and rupture [7].
- Imaging Technique: Use a custom-built Time-Domain Full-Field Optical Coherence Tomography (FF-OCT) system.
- Principle: FF-OCT is an interferometric technique that uses a broadband halogen light source to achieve sub-micrometer axial resolution. It enables scan-free, en face visualization of cellular structures in 3D without labels [7].
- Procedure: Initiate FF-OCT imaging immediately after drug administration. Acquire images continuously at 20-minute intervals for up to 180 minutes. Use a Linnik-configured Michelson interferometer with identical 40× water-immersion objectives for subcellular resolution. Reconstruct 3D surface topography from the acquired z-stacks [7].
- Key Observable Morphological Parameters [7]:
- For Apoptosis: Look for sequential appearance of echinoid spine formation, cell contraction, membrane blebbing, and filopodia reorganization.
- For Necrosis: Look for rapid membrane rupture, intracellular content leakage, and abrupt loss of adhesion structures.
Biochemical and Molecular Assays
While morphology is indicative, confirmation through biochemical assays is essential.
- Apoptosis Detection:
- Caspase-3/8 Cleavage: Detect cleaved (activated) caspase-3 or caspase-8 via western blotting or immunofluorescence. This is a gold-standard biomarker for apoptosis [3] [106].
- Phosphatidylserine (PS) Exposure: Use Annexin V staining, which binds to PS translocated to the outer leaflet of the plasma membrane. This is typically performed in conjunction with a viability dye (e.g., Propidium Iodide, PI) to distinguish early apoptotic (Annexin V+/PI-) from late apoptotic/necrotic (Annexin V+/PI+) cells [3] [106].
- Necroptosis Detection:
- Phospho-MLKL Detection: The phosphorylation of MLKL, particularly by RIPK3, is a definitive marker for necroptosis. This can be detected using phospho-specific antibodies in western blotting or immunofluorescence [108] [34].
- Inhibition Assays: Use specific inhibitors of RIPK1 (e.g., Necrostatin-1) or MLKL to confirm the involvement of the necroptotic pathway. A reduction in cell death upon inhibitor treatment supports the occurrence of necroptosis [108].
- General Cell Death and Inflammation Assessment:
- Lactate Dehydrogenase (LDH) Release Assay: Measure the release of LDH, a stable cytosolic enzyme, into the cell culture supernatant. High LDH release indicates loss of membrane integrity, a hallmark of lytic cell death like necrosis and necroptosis [102].
- Cytokine Release: Quantify the release of pro-inflammatory cytokines such as IL-1β and IL-18, or HMGB1 (a DAMP) to assess the immunogenic potential of the cell death modality [34] [102].
The Scientist's Toolkit: Essential Research Reagents
The table below lists key reagents and tools used in the featured experiment and for general research into these cell death pathways.
Table 2: Key Research Reagents for Cell Death Studies
| Reagent / Tool | Function/Application | Example Use Case |
|---|---|---|
| Doxorubicin | Inducer of DNA damage and intrinsic apoptosis [7] | Triggering apoptotic pathway in cancer cell lines like HeLa for experimental study [7]. |
| Ethanol (High Conc.) | Chemical inducer of necrosis; disrupts membrane integrity and denatures proteins [7] | Serving as a positive control for necrotic cell death in experimental models [7]. |
| TNF-α | Cytokine that activates TNFR1; can induce apoptosis or necroptosis depending on caspase activity [108] [102] | Studying death receptor-mediated pathways and the switch between apoptosis and necroptosis [34]. |
| Z-VAD-FMK | Pan-caspase inhibitor [34] | Blocking apoptotic pathways to shift cell fate towards caspase-independent death like necroptosis [34]. |
| Necrostatin-1 | Specific inhibitor of RIPK1 kinase activity [108] | Confirming RIPK1-dependent necroptosis in mechanistic studies [108]. |
| Anti-Cleaved Caspase-3 Antibody | Detects activated caspase-3 via WB, IHC, or IF [3] [106] | Gold-standard biomarker for confirming apoptosis execution [3]. |
| Anti-Phospho-MLKL Antibody | Detects phosphorylated MLKL via WB or IF [108] [34] | Definitive marker for ongoing necroptosis [108]. |
| Annexin V Conjugates | Binds to externalized PS for flow cytometry or microscopy [106] | Identifying early-stage apoptotic cells. |
| Propidium Iodide (PI) | DNA dye that is impermeable to live and early apoptotic cells [106] [102] | Distinguishing viable (PI-) from dead cells with compromised membranes (PI+). |
| CY-09 | Specific inhibitor of the NLRP3 inflammasome [102] | Investigating the role of pyroptosis and its crosstalk with other death pathways (e.g., in PANoptosis) [102]. |
Crosstalk and Integrated View: PANoptosis
Emerging evidence reveals significant crosstalk between apoptosis, necroptosis, and pyroptosis (an inflammatory form of death mediated by gasdermin proteins). This has led to the concept of PANoptosis, defined as a unique inflammatory programmed cell death pathway that is regulated by integrated signaling from all three mechanisms and cannot be fully accounted for by any of them alone [109] [102].
PANoptosis is characterized by the concurrent activation of key molecules from these pathways within a single cell, driven by a multiprotein complex called the PANoptosome [109] [102]. This phenomenon has been observed in response to specific triggers, including viral and bacterial infections, as well as in the context of cytokine storms and cancer [109]. For instance, a 2025 study demonstrated that TNF-α could induce PANoptosis in osteoblasts, contributing to the inhibition of osteogenic differentiation in inflammatory bone diseases [102]. Scanning electron microscopy revealed multiple PANoptosis morphologies in the same field of view, and inhibition of the NLRP3 inflammasome component could rescue cells from this combined death, highlighting the integrated nature of the process [102]. Understanding PANoptosis provides a more holistic framework for developing therapies for complex diseases where multiple cell death pathways are engaged simultaneously.
Using Pharmacological Inhibitors to Validate Molecular Pathways (e.g., Caspase, RIPK1)
The precise differentiation between apoptosis and necrosis is a cornerstone of modern cell biology, with profound implications for understanding disease mechanisms and developing targeted therapies. For researchers and drug development professionals, pharmacological inhibitors serve as indispensable tools for dissecting complex molecular pathways and validating the morphological hallmarks specific to each form of cell death. Apoptosis, characterized by cell shrinkage, chromatin condensation, and formation of apoptotic bodies, represents a caspase-dependent, programmed process that minimizes inflammatory response [63] [3]. In contrast, necroptosis—a regulated form of necrosis—presents with cell swelling, plasma membrane rupture, and release of pro-inflammatory intracellular contents, mediated through receptor-interacting serine/threonine protein kinase 1 (RIPK1), RIPK3, and mixed-lineage kinase domain-like protein (MLKL) [112] [3].
The strategic application of pathway-specific inhibitors allows researchers to experimentally manipulate these cell death modalities, establishing causal relationships between molecular signaling and morphological outcomes. This guide provides a comparative analysis of pharmacological inhibitors targeting key components of cell death pathways, with particular emphasis on RIPK1 and caspase inhibitors, to facilitate informed reagent selection and experimental design for validating morphological specificity in apoptosis versus necrosis research.
Molecular Mechanisms and Morphological Correlates
Apoptosis: The Caspase-Dependent Pathway
The extrinsic apoptosis pathway initiates when death ligands such as tumor necrosis factor-alpha (TNF-α) bind to cell surface receptors, leading to the formation of a death-inducing signaling complex (DISC) containing Fas-associated death domain (FADD) and pro-caspase-8 [113] [3]. Activated caspase-8 initiates a proteolytic cascade that executes the apoptotic program through effector caspases (caspase-3, -6, -7), resulting in characteristic morphological changes including cell shrinkage, membrane blebbing, chromatin condensation, and DNA fragmentation [113] [6]. The intrinsic apoptosis pathway triggers mitochondrial outer membrane permeabilization (MOMP), leading to cytochrome c release and activation of the caspase cascade via the apoptosome [113].
Necroptosis: The RIPK1/RIPK3/MLKL Axis
Necroptosis represents a caspase-independent, regulated form of cell death with necrotic morphology. Upon TNF receptor activation, RIPK1 is recruited to complex I alongside TRADD, TRAF2, and cellular inhibitor of apoptosis proteins (cIAP1/2) [112] [114]. When caspase-8 activity is compromised, RIPK1 interacts with RIPK3 through RIP homotypic interaction motifs (RHIM), forming the necrosome (complex IIb) [112]. This leads to RIPK3-mediated phosphorylation of MLKL, which oligomerizes, translocates to the plasma membrane, and causes membrane disruption, culminating in the release of damage-associated molecular patterns (DAMPs) and intense inflammation [112] [3].
Figure 1: Molecular switch between apoptotic and necroptotic pathways. Caspase-8 activity serves as the critical determinant directing cells toward apoptosis or necroptosis.
Comparative Analysis of Pharmacological Inhibitors
RIPK1 Inhibitors: Mechanisms and Applications
RIPK1 inhibitors represent a promising therapeutic strategy for inflammatory, neurodegenerative, and autoimmune conditions where necroptosis contributes to pathogenesis [114] [115]. These compounds target the kinase domain of RIPK1, preventing its activation and subsequent initiation of the necroptotic cascade.
Table 1: Comparative Analysis of RIPK1 Inhibitors
| Inhibitor | Mechanism/Target | Cellular IC₅₀ | Key Morphological Outcomes | Research Applications |
|---|---|---|---|---|
| Necrostatin-1 (Nec-1) | Type II ATP-competitive; binds DLG-out conformation [115] | ~0.3-0.5 μM [115] | Suppresses membrane rupture, cell swelling; preserves membrane integrity [112] | Experimental models of sepsis, ischemic injury [115] |
| Nec-1s (Stable Nec-1) | Enhanced stability version of Nec-1 [115] | ~0.1-0.3 μM [115] | Similar to Nec-1 with prolonged efficacy | Neurodegeneration models, inflammation studies |
| GSK'2982772 | Type II inhibitor; stabilizes inactive DLG-out conformation [116] | ~0.016-0.033 μM [116] | Prevents MLKL phosphorylation and membrane disruption | Phase II clinical trials for psoriasis, rheumatoid arthritis [116] |
| Phensuximide | FDA-approved anticonvulsant; RIPK1 inhibitor via structural similarity to Nec-1 [115] | ~40 μM [115] | Inhibits TSZ-induced necroptosis; reduces plasma membrane permeability | Drug repurposing for necroptosis-mediated conditions [115] |
| SKLB-Z30 | Novel scaffold; highly selective ATP-competitive inhibitor [116] | ~0.01 μM [116] | Blocks RIPK1/RIPK3/MLKL phosphorylation; attenuates cytokine release | Hemophagocytic lymphohistiocytosis (HLH) models [116] |
Caspase Inhibitors: Specificity and Limitations
Caspase inhibitors provide crucial tools for distinguishing caspase-dependent apoptosis from other cell death modalities. Pan-caspase inhibitors like z-VAD-fmk are frequently employed to experimentally induce a shift from apoptosis to necroptosis by blocking caspase-8 activity [112] [115].
Table 2: Caspase Inhibitors in Cell Death Differentiation
| Inhibitor | Specificity | Mechanism | Morphological Consequences | Experimental Considerations |
|---|---|---|---|---|
| z-VAD-fmk | Pan-caspase inhibitor [115] | Irreversible binding to catalytic site | Blocks apoptotic morphology; unmasks necroptosis in susceptible cells [112] | Can induce RIPK1-dependent apoptosis at high concentrations; may enhance inflammation |
| Emricasan | Pan-caspase inhibitor | Competitive substrate analog | Suppresses chromatin condensation and apoptotic body formation | Clinical testing in liver diseases; used to validate caspase-independent death |
| Q-VD-OPh | Broad-spectrum caspase inhibitor | Prevents caspase activation | More potent apoptosis inhibition than z-VAD; reduced off-target effects | Improved cellular permeability and stability over z-VAD |
| Ac-DEVD-CHO | Caspase-3/7 selective inhibitor | Blocks effector caspase activity | Precludes late apoptotic events (DNA fragmentation, PS externalization) | Confirms involvement of executioner caspases in death phenotype |
Experimental Protocols for Pathway Validation
Validating Necroptosis Inhibition: RIPK1-Targeting Approaches
Protocol 1: TSZ-Induced Necroptosis Inhibition Assay
Purpose: To evaluate RIPK1 inhibitor efficacy in preventing necroptosis induced by TNF-α/Smac mimetic/z-VAD (TSZ) treatment [115].
Reagents:
- TNF-α (10-50 ng/mL)
- Smac mimetic (e.g., BV6; 1-10 μM)
- z-VAD-fmk (10-50 μM)
- Test RIPK1 inhibitors (Nec-1, GSK'2982772, phensuximide, etc.)
- Cell viability assay (MTT, CellTiter-Glo, or propidium iodide exclusion)
- Phospho-specific antibodies (p-RIPK1 Ser166, p-MLKL Ser358)
Procedure:
- Seed HT-29 or L929 cells (commonly used necroptosis models) in 96-well plates (5,000-10,000 cells/well)
- Pre-treat cells with varying concentrations of RIPK1 inhibitors (0.1-100 μM) for 1-2 hours
- Induce necroptosis by adding TSZ cocktail: TNF-α (20 ng/mL) + Smac mimetic (5 μM) + z-VAD-fmk (20 μM)
- Incubate for 12-24 hours at 37°C, 5% CO₂
- Quantify cell death using viability assays and compare to inhibitor-free controls
- Confirm mechanism through Western blot analysis of RIPK1 and MLKL phosphorylation
- Assess morphological changes via microscopy: monitor membrane integrity, cell swelling, and formation of apoptotic bodies
Data Interpretation: Effective RIPK1 inhibitors demonstrate concentration-dependent protection against TSZ-induced death with IC₅₀ values typically in nanomolar to low micromolar range. Concurrent reduction in p-RIPK1 and p-MLKL confirms target engagement [115].
Protocol 2: LPS/TNF-Induced Systemic Inflammatory Response Syndrome (SIRS) Model
Purpose: To evaluate RIPK1 inhibitor efficacy in vivo using murine SIRS models relevant to sepsis pathogenesis [117] [115].
Reagents:
- Lipopolysaccharide (LPS; 10 mg/kg) or TNF-α (0.5 mg/kg)
- RIPK1 inhibitors (formulated for in vivo administration)
- Control compounds (vehicle, inactive analogs)
- Serum collection tubes for cytokine analysis
Procedure:
- Pre-treat mice (C57BL/6, 8-12 weeks) with RIPK1 inhibitors or vehicle (oral gavage or i.p. injection)
- After 1 hour, administer LPS or TNF-α intraperitoneally
- Monitor core body temperature every 2 hours for 12 hours using rectal thermometer
- Assess survival every 6 hours for 48-72 hours
- Collect serum at 6-hour post-challenge for cytokine profiling (TNF-α, IL-1β, IL-6) via ELISA
- For histopathological analysis, harvest tissues (lung, liver, spleen) for H&E staining and assessment of necroptotic morphology
Data Interpretation: Effective RIPK1 inhibitors significantly attenuate hypothermia, improve survival, reduce cytokine levels, and decrease tissue damage compared to vehicle controls [115].
Morphological Assessment Techniques
Light Microscopy: Employ hematoxylin and eosin (H&E) staining to distinguish apoptotic cells (shrunken with condensed, fragmented nuclei) from necroptotic cells (swollen with maintained nuclear structure until late stages) [6].
Fluorescence Microscopy: Utilize Hoechst 33342 or DAPI to visualize nuclear morphology. Apoptotic cells show brightly stained, condensed, and fragmented nuclei, while necroptotic cells maintain relatively normal nuclear morphology until membrane rupture [6].
Transmission Electron Microscopy (TEM): The gold standard for differentiating cell death modalities. Apoptosis displays chromatin margination, intact organelles, and apoptotic bodies. Necroptosis shows organelle swelling, plasma membrane rupture, and intact nuclear envelopes until late stages [6].
Membrane Integrity Assessment: Combine propidium iodide (PI) with Hoechst 33342. Necroptotic cells show PI positivity due to membrane disruption, while apoptotic cells exclude PI until secondary necrosis occurs [6].
Figure 2: Experimental workflow for validating cell death pathways using pharmacological inhibitors and morphological analysis.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagent Solutions for Cell Death Pathway Analysis
| Reagent Category | Specific Examples | Research Application | Experimental Considerations |
|---|---|---|---|
| RIPK1 Inhibitors | Nec-1, Nec-1s, GSK'2982772, phensuximide, SKLB-Z30 [115] [116] | Inhibit RIPK1 kinase activity to validate necroptosis involvement | Assess specificity using kinase panels; monitor potential off-target effects on apoptosis |
| Caspase Inhibitors | z-VAD-fmk (pan-caspase), Q-VD-OPh (broad-spectrum), Ac-DEVD-CHO (caspase-3/7) [112] [115] | Block apoptotic signaling to unmask alternative death pathways | z-VAD can promote necroptosis when caspase-8 is inhibited; use multiple inhibitors to confirm findings |
| Death Inducers | TNF-α (complex I/II formation), TSZ cocktail (necroptosis induction), Smac mimetics (cIAP depletion) [112] [115] | Activate specific cell death pathways in controlled experimental settings | Optimize concentrations for cell type; time-course experiments recommended |
| Antibodies | Phospho-RIPK1 (Ser166), phospho-MLKL (Ser358), cleaved caspase-3, caspase-8 [117] [115] | Detect pathway activation and inhibitor target engagement | Validate antibodies in relevant models; use multiple antibodies to confirm key pathway events |
| Viability Assays | MTT, CellTiter-Glo, propidium iodide, Sytox Green | Quantify cell death and inhibitor efficacy | Combine with morphological assessment; viability assays alone cannot differentiate death modalities |
| Cytokine ELISA | TNF-α, IL-1β, IL-6, IL-18 [117] | Measure inflammatory response associated with necroptosis | Particularly important for in vivo studies and immunogenic cell death assessment |
Clinical Translation and Therapeutic Applications
The therapeutic potential of RIPK1 inhibitors is currently being explored across multiple disease domains, with several candidates advancing to clinical trials. In sepsis and systemic inflammatory response syndrome (SIRS), RIPK1 inhibition has demonstrated significant protection in preclinical models. A clinical study measuring necroptosis-related proteins in ICU patients found that RIPK1, IL-1β, and IL-18 were significantly elevated in sepsis patients compared to controls, while caspase-8 levels were reduced—indicating a shift toward necroptotic cell death [117]. RIPK1 showed strong predictive value for sepsis/septic shock (AUC = 0.81), supporting its potential as both biomarker and therapeutic target [117].
Notably, drug repurposing approaches have identified FDA-approved compounds with RIPK1 inhibitory activity. Phensuximide, an anticonvulsant, has demonstrated efficacy in inhibiting RIPK1 kinase activity and protecting against LPS- and TNF-induced SIRS in murine models [115]. This strategy accelerates therapeutic development by leveraging existing safety profiles.
In autoimmune and neurodegenerative conditions, RIPK1 inhibitors have shown promise in preclinical models. DNL747 (SAR443820) advanced to Phase II trials for Alzheimer's disease and multiple sclerosis, while GDC-8264 is being evaluated for graft-versus-host disease [115]. These developments underscore the growing therapeutic relevance of targeted pathway inhibition in cell death-driven pathologies.
Pharmacological inhibitors provide indispensable tools for validating molecular pathways and establishing causal relationships between specific signaling events and morphological outcomes in cell death research. The strategic application of RIPK1 and caspase inhibitors enables researchers to dissect the complex interplay between apoptotic and necroptotic pathways, revealing context-dependent cellular decisions with significant implications for understanding disease mechanisms and developing targeted therapies. As inhibitor specificity and pharmacokinetic properties continue to advance, these compounds will play an increasingly important role in both basic research and translational medicine, particularly for inflammatory, neurodegenerative, and oncological conditions where regulated cell death contributes to pathogenesis.
Integrating Morphological Data with Flow Cytometry and Western Blot Analysis
In cell death research, distinguishing between apoptosis and necrosis is fundamental, with morphological assessment serving as the historical and foundational criterion. While biochemical assays like flow cytometry and Western blot provide precise molecular data, they are most powerful when correlated with the distinct morphological landscapes of different cell death pathways. Apoptosis is characterized by a controlled, energy-dependent process featuring cell shrinkage, chromatin condensation, nuclear fragmentation, and plasma membrane blebbing, ultimately resulting in the formation of apoptotic bodies that are neatly phagocytosed without inducing inflammation [3] [118]. In stark contrast, necrosis presents as an uncontrolled process involving cell swelling, plasma membrane rupture, and organelle breakdown, leading to the release of intracellular contents and a significant inflammatory response [3] [119]. This guide objectively compares the performance of morphological analysis, flow cytometry, and Western blotting in differentiating these pathways, providing integrated experimental protocols and data to empower researchers in making informed methodological choices.
Morphological Features: The Gold Standard for Classification
The initial classification of programmed cell death (PCD) into three distinct types—apoptosis (Type I), autophagic cell death (Type II), and necrosis (Type III)—was based primarily on morphological characteristics observed through microscopy [3]. These visual hallmarks remain the definitive standard for confirming the mode of cell death, providing context for molecular data obtained from other techniques.
Apoptotic cells undergo a characteristic sequence of structural changes. The cell shrinks and loses contact with its neighbors. The chromatin condenses and margins against the nuclear envelope (pyknosis), and the nucleus fragments (karyorrhexis). The plasma membrane forms blebs, eventually pinching off into membrane-enclosed apoptotic bodies containing intact organelles and nuclear fragments [3] [118]. Critically, the plasma membrane retains its integrity until the final stages, preventing inflammatory responses.
Necrotic cells follow a different path. They exhibit an early swelling of the cytoplasm and organelles, especially mitochondria. The plasma membrane loses its integrity, becomes ruptured, and leaks intracellular contents, a key event that promotes inflammation. The nucleus undergoes pyknosis and eventual dissolution (karyolysis), but without the formation of discrete apoptotic bodies [3] [119].
Table 1: Core Morphological Differences Between Apoptosis and Necrosis
| Cellular Feature | Apoptosis | Necrosis |
|---|---|---|
| Cell Size | Shrinkage | Swelling |
| Plasma Membrane | Blebbing, intact until late stages | Rapid rupture, loss of integrity |
| Nucleus | Chromatin condensation, nuclear fragmentation | Disintegration, karyolysis |
| Organelles | Largely intact until late stages | Gross swelling, dissolution |
| Inflammatory Response | None (phagocytic clearance) | Significant |
| Elimination | Phagocytosis by neighboring cells | Cell lysis in situ |
Advanced, label-free imaging technologies are now quantifying these morphological distinctions with high resolution. Full-field optical coherence tomography (FF-OCT) allows for the precise 3D visualization of apoptotic features like echinoid spine formation and membrane blebbing, versus necrotic membrane rupture and content leakage [7]. Furthermore, machine learning platforms such as LANCE (Live, Apoptotic, and Necrotic Cell Explorer) can automatically categorize cell death states from brightfield images with over 96% accuracy, providing a high-throughput, non-destructive method for morphological assessment [120].
Integrated Experimental Workflows and Protocols
A robust comparison of cell death pathways requires a multi-faceted approach. Below are detailed protocols for inducing cell death and for analyzing the outcomes using morphology, flow cytometry, and Western blotting in an integrated manner.
Cell Death Induction and Sample Preparation
A. Induction of Apoptosis
- Reagents: Tumor Necrosis Factor-alpha (TNF-α) (e.g., 20 ng/mL), Cycloheximide (CHX) (e.g., 20 μg/mL), Camptothecin (1-10 μM), or anti-Fas/CD95 antibodies (e.g., clone Jo2) [118] [120].
- Procedure: Seed cells (e.g., U937, Jurkat) and allow to adhere. Replace medium with fresh medium containing the apoptosis-inducing agent. Incubate for a predetermined time (e.g., 4-6 hours). Harvest both adherent and floating cells for analysis [120].
B. Induction of Necrosis/Necroptosis
- Reagents: High-dose Hydrogen Peroxide (H₂O₂) (e.g., 2 mM), or a combination of TNF-α (20 ng/mL) + Cycloheximide (20 μg/mL) + pan-caspase inhibitor z-VAD-fmk (e.g., 100 μM) [121] [120].
- Procedure: Seed cells as above. For chemical induction, treat with H₂O₂ for several hours. For necroptosis, pre-treat with z-VAD-fmk for 30-60 minutes before adding TNF-α and CHX. Incubate for 6-8 hours and harvest all cells [120].
Key Analysis Workflow
The following workflow diagram illustrates how these techniques can be combined in a single experiment.
Diagram Title: Integrated Cell Death Analysis Workflow
Comparative Performance of Detection Methods
Flow Cytometry Assays
Flow cytometry offers high-throughput, quantitative data on cell populations. The Annexin V/PI assay is a cornerstone for distinguishing early apoptosis from late apoptosis/necrosis.
Detailed Annexin V/PI Protocol [122] [118]:
- Harvest and Wash: Harvest induced and control cells. Wash cells twice with cold phosphate-buffered saline (PBS).
- Resuspend in Binding Buffer: Resuspend approximately 1x10⁵ cells in 100-400 μL of 1X Annexin V Binding Buffer.
- Staining: Add 5 μL of Fluorochrome-conjugated Annexin V (e.g., FITC, PE) and 10 μL of Propidium Iodide (PI) working solution (or 7-AAD) to the cell suspension.
- Incubation: Gently vortex the cells and incubate for 15 minutes at room temperature (25°C) in the dark.
- Analysis: Within 1 hour, analyze the cells by flow cytometry. Use FITC (or equivalent) and PI channels.
- Annexin V-/PI-: Viable, non-apoptotic cells.
- Annexin V+/PI-: Early apoptotic cells (PS externalized, membrane intact).
- Annexin V+/PI+: Late apoptotic or necroptotic cells.
- Annexin V-/PI+: Necrotic cells or cellular debris.
Other vital flow cytometry assays include:
- Caspase Activation: Using cell-permeable FLICA (Fluorochrome-Labeled Inhibitors of Caspases) probes or antibodies specific to the active cleaved form of caspases (e.g., caspase-3) [122] [118].
- Mitochondrial Membrane Potential (ΔΨm): Using cationic dyes like TMRM or JC-1, which accumulate in active mitochondria; a loss of fluorescence indicates ΔΨm dissipation, an early apoptotic event [122].
- DNA Fragmentation (TUNEL Assay): Detects endonucleolytic DNA cleavage by labeling DNA strand breaks with Br-dUTP, which is then detected by a fluorescent anti-BrdU antibody [118].
Table 2: Flow Cytometry Assays for Cell Death Analysis
| Assay | Target | Apoptosis Signature | Necrosis Signature | Key Advantage |
|---|---|---|---|---|
| Annexin V/PI | PS exposure & membrane integrity | Annexin V+/PI- (early) | Annexin V+/PI+ or Annexin V-/PI+ | Gold standard for early apoptosis |
| Active Caspase-3 | Caspase-3 cleavage | High positive population | Low/No positive population | Specific for core apoptotic machinery |
| TMRM/JC-1 | Mitochondrial potential (ΔΨm) | Loss of fluorescence | Variable, often late loss | Detects early intrinsic pathway event |
| TUNEL | DNA fragmentation | High positive population | Low/No positive population | Detects late, irreversible stage |
Western Blot Analysis
Western blotting provides molecular-level confirmation by detecting key protein markers and their activation states.
Detailed Western Blot Protocol [123] [102]:
- Protein Extraction: Lyse harvested cells in RIPA buffer supplemented with protease and phosphatase inhibitors on ice for 30 minutes. Centrifuge at 12,000-14,000 rpm for 15 minutes at 4°C to collect the supernatant.
- Protein Quantification: Determine protein concentration using a BCA assay. Normalize all samples to the same concentration.
- Gel Electrophoresis: Load 20-40 μg of protein per lane on an SDS-PAGE gel and run at constant voltage until the dye front reaches the bottom.
- Membrane Transfer: Transfer proteins from the gel to a PVDF or nitrocellulose membrane using a wet or semi-dry transfer system.
- Blocking and Antibody Incubation: Block the membrane with 5% non-fat milk or BSA in TBST for 1 hour. Incubate with primary antibodies diluted in blocking buffer overnight at 4°C. Key antibodies include:
- Anti-Cleaved Caspase-3: Appears as bands at ~17/19 kDa.
- Anti-PARP: Full-length (116 kDa) and cleaved fragment (89 kDa).
- Anti-Bax / Anti-Bcl-2: To assess pro- vs. anti-apoptotic balance.
- Anti-LC3B / Anti-Beclin-1: For autophagy detection.
- Anti-β-Actin / Anti-GAPDH: Loading control.
- Detection: Wash membrane and incubate with an HRP-conjugated secondary antibody. Develop using an ECL reagent and visualize with a chemiluminescence imager.
The signaling pathways governing apoptosis and necrosis involve distinct molecular players, which can be visualized as follows.
Diagram Title: Apoptosis and Necroptosis Signaling Pathways
Table 3: Key Protein Biomarkers for Western Blot Analysis
| Biomarker | Molecular Weight (Full-length/Cleaved) | Role in Apoptosis | Role in Necrosis/Necroptosis |
|---|---|---|---|
| Caspase-3 | 32 kDa / 17, 19 kDa | Cleavage and activation is a central execution step | Not activated (in necroptosis) |
| PARP | 116 kDa / 89 kDa | Cleaved by caspase-3, inactivating DNA repair | Not cleaved (in primary necrosis) |
| Bax/Bcl-2 Ratio | 21 kDa / 26 kDa | Ratio increases, promoting mitochondrial leakage | Unchanged or variable |
| LC3B | 16, 18 kDa (I/II) | May be upregulated; indicates autophagy | Not typically associated |
| p-MLKL | 54 kDa | Not phosphorylated | Phosphorylation is a key necroptosis marker |
Research Reagent Solutions
A successful integrated analysis relies on a toolkit of well-validated reagents. The table below lists essential materials and their functions.
Table 4: Essential Reagents for Cell Death Analysis
| Reagent / Kit | Primary Function | Key Application |
|---|---|---|
| Fluorochrome-labeled Annexin V | Binds externalized phosphatidylserine (PS) | Flow cytometry: detection of early apoptosis |
| Propidium Iodide (PI) / 7-AAD | Nucleic acid intercalation in membrane-compromised cells | Flow cytometry: distinguishes viable from dead cells |
| FLICA Probes | Irreversibly binds active caspase enzymes | Flow cytometry/Live-cell imaging: caspase activity |
| Antibody: Cleaved Caspase-3 | Recognizes activated caspase-3 fragment | Western Blot/Flow cytometry: confirmation of apoptosis |
| Antibody: Cleaved PARP | Recognizes caspase-cleaved PARP fragment | Western Blot: specific apoptosis biomarker |
| Antibody: Phospho-MLKL | Recognizes activated MLKL | Western Blot: specific necroptosis biomarker |
| TMRM / JC-1 Dye | Accumulates in polarized mitochondria | Flow cytometry: measurement of mitochondrial potential (ΔΨm) |
| LDH Release Assay Kit | Measures Lactate Dehydrogenase enzyme in supernatant | Spectrophotometry: quantifies cytosolic leakage in necrosis |
| Cell Viability Stains (Calcein AM) | Live-cell esterase activity probe | Fluorescence microscopy: identifies viable cells |
| Nucleic Acid Stains (Hoechst) | Binds DNA, shows chromatin condensation | Fluorescence microscopy: visualizes nuclear morphology |
Comparative Morphology in Tissue Context vs. Cell Culture
The accurate identification of cell death modalities is fundamental to biomedical research, influencing everything from our understanding of basic physiology to the development of novel therapeutics. For decades, the binary distinction between apoptosis and necrosis has served as a cornerstone of pathological assessment. Apoptosis is a genetically regulated, energy-dependent process characterized by specific morphological and biochemical changes that allow for the organized disposal of cells without inciting an inflammatory response. In contrast, necrosis has traditionally been viewed as an accidental, unregulated form of cell death resulting from overwhelming physicochemical insult, leading to plasma membrane rupture and the release of intracellular contents that provoke inflammation [63] [10]. However, the discovery of regulated necrotic pathways, such as necroptosis, has blurred this simple dichotomy, revealing that some forms of necrosis are also subject to molecular regulation [3] [10].
Within this evolving paradigm, the morphological features of dying cells remain primary diagnostic criteria. Yet, the manifestation of these features can vary significantly depending on the physiological context—whether in a complex, multi-cellular tissue environment or in a controlled cell culture system. This guide provides a detailed comparison of the morphological features of apoptosis and necrosis as they appear in these two distinct contexts, equipping researchers with the knowledge to accurately interpret cell death phenomena across experimental systems.
Core Morphological Definitions of Apoptosis and Necrosis
The foundational morphological characteristics of apoptosis and necrosis were first systematically defined by Kerr, Wyllie, and Currie in 1972 [10]. These criteria, established through histological and electron microscopic examination of tissues, remain the gold standard for differentiation.
Characteristic Features of Apoptosis
Apoptosis is an active, programmed process of autonomous cellular dismantling. Its key morphological features include:
- Cell Shrinkage and Cytoplasmic Condensation: The cell undergoes a reduction in volume, with the cytoplasm becoming denser [63] [10].
- Nuclear Fragmentation (Karyorrhexis): The chromatin condenses intensely and margins under the nuclear membrane, a stage known as pyknosis. The nucleus then breaks into discrete fragments [3] [63].
- Plasma Membrane Blebbing: The cell surface forms protrusions, a process driven by caspase-mediated activation of gelsolin and cleavage of focal adhesion kinases [63].
- Formation of Apoptotic Bodies: The cell fragments into multiple, membrane-bound vesicles containing intact organelles and nuclear fragments [3] [10].
- Phagocytosis: Apoptotic bodies are swiftly engulfed by neighboring phagocytic cells (e.g., macrophages or parenchymal cells), preventing the release of cellular contents and avoiding an inflammatory response [63] [10].
Characteristic Features of Necrosis
Necrosis is characterized by a loss of regulatory control and catastrophic cell failure.
- Cell and Organelle Swelling (Oncosis): One of the earliest signs is an increase in cell volume due to the failure of ion pumps and water influx [3] [17].
- Plasma Membrane Rupture: The loss of membrane integrity leads to the uncontrolled release of intracellular components into the extracellular space [7] [10].
- Organelle Degradation: Mitochondria and other organelles swell and undergo disintegration [3].
- Nuclear Changes: The nucleus may undergo pyknosis, karyorrhexis, and finally karyolysis, where the nuclear chromatin dissolves due to enzymatic degradation [3] [10].
- Inflammatory Response: The leakage of intracellular molecules, known as Damage-Associated Molecular Patterns (DAMPs), triggers a potent inflammatory reaction in the surrounding tissue [63] [10].
Table 1: Core Morphological Features of Apoptosis and Necrosis
| Feature | Apoptosis | Necrosis (Incl. Necroptosis) |
|---|---|---|
| Cell Size | Shrinkage | Swelling (Oncosis) |
| Plasma Membrane | Intact, but blebbing; Phosphatidylserine exposure | Ruptured, loss of integrity |
| Nucleus | Chromatin condensation, fragmentation (Karyorrhexis) | Karyolysis (dissolution) or irregular fragmentation |
| Organelles | Initially intact, packaged into apoptotic bodies | Swollen, degraded |
| Cellular Contents | Retained in apoptotic bodies | Leaked into extracellular space |
| Fate of Dead Cell | Phagocytosed by adjacent cells | In situ dissolution |
| Inflammatory Response | No | Yes |
Morphology in Tissue Context
In a living tissue, cell death occurs within a structured, multi-cellular environment that actively participates in and responds to the process. This context profoundly influences the morphological presentation.
Tissue-Specific Dynamics and Challenges
Within tissues, apoptosis is a rapid and efficient mechanism for cell removal. The swift phagocytosis of apoptotic bodies by neighboring cells means that apoptotic cells are often visible only transiently in histological sections [17]. This can lead to an underestimation of their frequency. A key challenge in tissue analysis is the presence of TUNEL-positive staining in necrotic cells, a finding often misinterpreted as specific for apoptosis. As noted in a study of hepatic ischemia-reperfusion injury, "most necrotic cells stained positive with the TUNEL assay," underscoring that this assay detects DNA strand breaks common to both death modalities and is not specific for apoptosis [17]. Therefore, morphological assessment by a trained pathologist remains crucial for definitive classification.
Visualizing Cell Death Morphology in Tissues
The diagram below illustrates the key morphological differences and tissue-level consequences of apoptosis and necrosis.
Morphology in Cell Culture
Cell culture systems offer a simplified and controlled environment for studying cell death, but this very simplicity can alter the morphological progression and interpretation of death pathways.
Altered Dynamics in an Isolated System
The most significant difference in cell culture is the absence of a professional phagocytic system. An apoptotic cell cannot be cleared by a neighbor. Consequently, it progresses to a stage known as secondary necrosis [52] [124]. In this stage, the apoptotic cell, having undergone shrinkage and fragmentation, begins to lose its membrane integrity, becoming permeable to dyes like propidium iodide (PI). This makes it morphologically and biochemically difficult to distinguish from a primary necrotic cell, especially at late time points [52] [124]. This is a major confounding factor in quantitative assays.
High-Resolution Imaging of Single-Cell Death
Advanced label-free imaging techniques like Full-Field Optical Coherence Tomography (FF-OCT) have allowed for detailed, real-time observation of death in single cells. Studies using FF-OCT have captured the dynamic sequence of apoptosis induced by doxorubicin, showing "characteristic features such as echinoid spine formation, cell contraction, membrane blebbing, and filopodia reorganization" [7]. In contrast, ethanol-induced necrosis was marked by "rapid membrane rupture, intracellular content leakage, and abrupt loss of adhesion structure" without the organized blebbing seen in apoptosis [7].
Visualizing Cell Death Progression in Culture
The following diagram outlines the distinct and overlapping pathways of cell death in a culture system, highlighting the critical juncture of secondary necrosis.
Direct Comparative Analysis: Tissue vs. Cell Culture
The following table synthesizes the key morphological differences for apoptosis and necrosis when observed in their native tissue context versus the artificial environment of cell culture.
Table 2: Morphological Comparison of Cell Death in Tissue vs. Cell Culture
| Aspect | Tissue Context | Cell Culture Context |
|---|---|---|
| Apoptosis: Fate of Cell Corpse | Rapid phagocytosis by adjacent cells or professional phagocytes. Apoptotic bodies are transient. | No phagocytic system. Apoptotic bodies remain and progress to secondary necrosis. |
| Apoptosis: Inflammatory Outcome | None (immunologically silent). | Potentially pro-inflammatory due to secondary necrosis and content release. |
| Necrosis: Tissue Response | Robust sterile inflammatory response; DAMPs recruit immune cells. | Inflammatory signals are released into the medium, but without a responsive tissue infrastructure. |
| Key Analytical Challenge | Distinguishing true apoptosis from other TUNEL-positive processes (e.g., necroptosis, oncosis). | Distinguishing late/secondary apoptotic cells from primary necrotic cells. |
| Optimal Identification Method | Histological evaluation by a pathologist (H&E staining), combined with specific immunohistochemistry for biomarkers (e.g., cleaved caspase-3). | Multi-parameter assays combining flow cytometry (Annexin V/PI) with imaging of nuclear morphology [124]. |
Experimental Protocols for Distinction
Accurately differentiating between apoptosis and necrosis, particularly in cell culture, requires a combination of techniques.
Imaging Flow Cytometry with Annexin V/PI Staining
This protocol allows for the quantitative distinction between apoptosis and necroptosis at the single-cell level by combining classical staining with image-based nuclear analysis [124].
- Key Reagents: Annexin V-FLUOS, Propidium Iodide (PI), caspase inhibitor (zVAD-fmk), IAP antagonist (BV6 for necroptosis induction).
- Methodology:
- Induce apoptosis (e.g., with CD95L) or necroptosis (CD95L + zVAD-fmk + BV6) in cells.
- Harvest and stain cells with Annexin V and PI.
- Analyze using an imaging flow cytometer (e.g., FlowSight).
- Discrimination Criteria: Apoptotic cells are Annexin V+/PI- (early) or Annexin V+/PI+ with nuclear fragmentation. Necroptotic cells are Annexin V+/PI+ but display nuclear swelling instead of fragmentation [124]. This direct visualization of nuclear morphology is key to overcoming the ambiguity of late-stage culture death.
Label-Free Live-Cell Imaging with FF-OCT
This technique enables non-invasive, high-resolution 3D monitoring of unlabeled cells, capturing dynamic morphological changes in real time [7].
- Methodology:
- Culture cells on an imaging dish.
- Treat with apoptosis inducer (e.g., 5 μmol/L doxorubicin) or necrosis inducer (e.g., 99% ethanol).
- Place the dish on a custom-built time-domain FF-OCT system immediately after treatment.
- Acquire images continuously at set intervals (e.g., every 20 min for up to 180 min).
- Data Analysis: Reconstruct 3D surface topography and tomography to quantify changes in cell volume, membrane dynamics (blebbing vs. rupture), and adhesion structures.
The Scientist's Toolkit: Key Reagents and Assays
Table 3: Essential Reagents and Tools for Cell Death Analysis
| Reagent/Assay | Function/Principle | Application Context |
|---|---|---|
| Annexin V | Binds to phosphatidylserine (PS) exposed on the outer leaflet of the plasma membrane in early apoptosis. | Flow Cytometry, Microscopy (Cell Culture) |
| Propidium Iodide (PI) | A membrane-impermeant dye that stains DNA in cells with compromised plasma membranes (necrotic and late apoptotic). | Flow Cytometry (Cell Culture) |
| Caspase Inhibitors (e.g., zVAD-fmk) | Pan-caspase inhibitor; inhibits apoptosis. Used to confirm caspase-dependence or to shift death to necroptosis. | Cell Culture (Mechanistic Studies) |
| TUNEL Assay | Detects DNA strand breaks. Labels apoptotic nuclei but is not specific; also stains necrotic cells. | Histology (Tissue), Cell Culture |
| Antibody vs. Cleaved Caspase-3 | Detects the activated form of the key executioner caspase, a specific biomarker for apoptosis. | Western Blot, IHC (Tissue & Culture) |
| Biomarkers (HMGB1, miR-122) | HMGB1 (hyper-acetylated form indicates immune activation) and miR-122 are released from necrotic cells. | Plasma/Serum Analysis (in vivo models) |
The targeted induction of cell death is a cornerstone of cancer treatment, with most chemotherapeutic agents designed to exploit specific molecular vulnerabilities in cancer cells to trigger their demise. The efficacy and toxicity profiles of these drugs are largely determined by the particular cell death pathway they activate. Historically, the paradigm of cell death was dichotomized into apoptosis and necrosis; however, advancements in cell biology have revealed a more complex landscape of regulated cell death (RCD) pathways, including necroptosis, pyroptosis, and ferroptosis [125]. Understanding the precise mechanisms by which chemotherapeutic agents preferentially activate one pathway over another is not only crucial for understanding their mechanism of action but also for overcoming drug resistance and minimizing off-target effects. This guide provides a comparative analysis of how different classes of chemotherapeutic drugs induce specific types of cell death, framed within the critical context of morphological and biochemical specificity.
The selectivity of cell death induction hinges on the drug's primary molecular target and the unique cellular context of the tumor. For instance, agents that cause DNA damage often engage the intrinsic apoptotic pathway, while death receptor agonists can directly trigger extrinsic apoptosis [126] [127]. Meanwhile, emerging evidence indicates that certain drugs, under specific conditions such as caspase inhibition, can shift the cell death modality to necroptosis or other alternative pathways [127] [128]. This comparative analysis will delve into the experimental data and methodologies used to decipher these death signals, providing researchers with a structured framework for evaluating the mechanistic actions of chemotherapeutic agents.
Comparative Analysis of Cell Death Pathways
Defining Morphological and Molecular Hallmarks
The classification of cell death types initially relies on distinct morphological features, which are a direct consequence of underlying molecular events. Apoptosis, often termed "programmed cell death," is characterized by cell shrinkage, chromatin condensation, nuclear fragmentation, and formation of apoptotic bodies that are neatly phagocytosed by neighboring cells, preventing an inflammatory response [63] [129] [128]. In stark contrast, necrosis involves cell and organelle swelling, loss of plasma membrane integrity, and uncontrolled release of cellular contents, which frequently provokes a robust inflammatory reaction [129] [125]. Necroptosis, a regulated form of necrosis, shares the lytic features of necrosis but is molecularly controlled [128]. Ferroptosis is defined by iron-dependent lipid peroxidation, leading to mitochondrial shrinkage and increased membrane density [130].
Table 1: Fundamental Characteristics of Major Cell Death Pathways
| Feature | Apoptosis | Necrosis | Necroptosis | Ferroptosis |
|---|---|---|---|---|
| Regulation | Programmed (Tightly Regulated) | Accidental (Unregulated) | Programmed | Programmed |
| Primary Inducers | Death ligands, DNA damage, ER stress [125] | Extreme physical/chemical injury [129] | TNFα, TLR ligands (when caspases are inhibited) [128] | GPX4 inhibition, iron overload, ROS [130] |
| Key Biochemical Mediators | Caspases (e.g., Caspase-3, -8, -9), Bcl-2 family [63] [125] | N/A (Unregulated) | RIPK1, RIPK3, MLKL [131] [128] | Iron, Lipid ROS, GPX4 loss [130] |
| Membrane Integrity | Maintained until late stages (blebbing) [129] | Lost early (lysis) [129] | Lost (lysis) [128] | Lost (permeabilization) |
| Inflammatory Response | Typically non-inflammatory [128] | Strongly inflammatory [129] | Immunogenic [128] | Immunogenic [130] |
| Morphology of Mitochondria | Condensation, cytochrome c release [125] | Swelling, fragmentation [128] | Swelling | Shrinkage, increased membrane density [130] |
Chemotherapeutic Agents and Their Preferential Death Pathways
Different classes of chemotherapeutic drugs have been shown to preferentially induce specific types of cell death based on their mechanism of action. The response can also be influenced by the cellular context, including the expression of key regulatory proteins.
Table 2: Preferential Cell Death Induction by Representative Chemotherapeutic Agents
| Drug Class / Agent | Primary Molecular Target | Preferential Cell Death Pathway | Key Experimental Evidence |
|---|---|---|---|
| Topoisomerase Inhibitors (e.g., Etoposide, Camptothecin) | Topoisomerase I/II, causing DSBs and DPCs [126] | Apoptosis (Intrinsic Pathway) | Induces mitochondrial outer membrane permeabilization (MOMP), caspase-3 activation, and phosphatidylserine externalization [126] [127]. |
| Platinum-based Agents (e.g., Cisplatin) | DNA (forming intrastrand cross-links, ICLs, and DPCs) [126] | Apoptosis (Intrinsic Pathway) | Triggers DNA damage response, p53 activation, and caspase-9-mediated apoptosis [126] [127]. Can also induce necroptosis in some settings [127]. |
| Alkylating Agents (e.g., Melphalan - L-PAM) | DNA (forming DSBs, DPCs, ICLs) [126] | Apoptosis | Annexin-V positivity and caspase dependency demonstrated in HeLa cells [126]. |
| TRAIL / Agonistic Antibodies | Death Receptors DR4/DR5 [132] | Apoptosis (Extrinsic Pathway) | Selective induction of apoptosis via formation of the Death-Inducing Signaling Complex (DISC) and activation of caspase-8 [132]. |
| DNMT Inhibitors (e.g., AzadC) | DNA methyltransferases (trapping, forming DPCs) [126] | Apoptosis | Shows significant Annexin-V binding in cell-based assays, indicating apoptosis [126]. |
| PARP Inhibitors (e.g., Olaparib, Niraparib) | PARP enzyme, DNA repair [130] | Apoptosis & Ferroptosis | Induces synthetic lethality in HRD cells (apoptosis). Can also induce ferroptosis via p53-dependent SLC7A11 downregulation or CD36-mediated lipid peroxidation [130]. |
| Anthracyclines (e.g., Doxorubicin) | DNA intercalation, Topoisomerase II inhibition [127] | Apoptosis, Necroptosis, Pyroptosis | Can trigger multiple pathways; apoptosis is primary, but necroptosis and pyroptosis have been observed in various models, contributing to efficacy and side effects [127]. |
Experimental Protocols for Cell Death Identification
Core Methodologies for Discriminating Death Pathways
A combination of techniques is required to definitively characterize the type of cell death induced by a chemotherapeutic agent. The following protocols represent standard methodologies cited in the literature.
Protocol 1: Annexin V/Propidium Iodide (PI) Staining for Apoptosis and Necrosis This flow cytometry-based assay is a cornerstone for distinguishing early apoptosis, late apoptosis, and necrosis.
- Procedure: Cells are harvested after drug treatment and stained with FITC-conjugated Annexin V and PI.
- Principle: Annexin V binds to phosphatidylserine (PS), which is externalized to the outer leaflet of the plasma membrane during early apoptosis. PI is a DNA dye that is excluded from live and early apoptotic cells but enters cells upon loss of membrane integrity, a hallmark of necrosis and late-stage apoptosis.
- Interpretation: Annexin V+/PI- cells are in early apoptosis; Annexin V+/PI+ cells are in late apoptosis or undergoing necrosis; Annexin V-/PI+ cells are considered necrotic [126]. This method was used to demonstrate apoptosis induction by topoisomerase inhibitors and AzadC [126].
Protocol 2: Caspase Activity Assay Confirming the involvement of caspases is critical for verifying apoptotic death.
- Procedure: Cell lysates from treated samples are incubated with caspase-specific substrates (e.g., DEVD for caspase-3, IETD for caspase-8, LEHD for caspase-9). Substrate cleavage releases a fluorescent or colorimetric signal.
- Principle: Activated caspases cleave these synthetic substrates after specific aspartic acid residues.
- Interpretation: An increase in signal indicates caspase activation. Pan-caspase inhibitors like zVAD-FMK can be used to confirm caspase dependency. If cell death is inhibited by zVAD, it is likely apoptotic; if not, an alternative pathway like necroptosis may be involved [131] [127].
Protocol 3: Western Blot Analysis for Pathway-Specific Markers This method detects molecular signatures of different death pathways.
- Key Targets:
- Application: Used to demonstrate that enalapril, zVAD-FMK, Necrostatin-1, and Ferrostatin-1 reduce cleaved caspase-3 and pMLKL levels in post-MI rats, confirming the inhibition of apoptosis and necroptosis, respectively [131].
Protocol 4: Clonogenic Survival Assay This gold-standard assay measures the long-term reproductive viability of cells after drug treatment, reflecting the ultimate cytotoxic effect regardless of the initial death modality.
- Procedure: Cells are plated at low density, treated with the chemotherapeutic agent, and then allowed to grow in drug-free medium for 7-14 days to form colonies. Colonies are stained and counted.
- Interpretation: The surviving fraction is calculated relative to untreated controls. This method provides a direct measure of a drug's capacity to induce irreversible cell death (of any type) and prevent proliferation, as employed in the GDSC drug sensitivity database [133].
Visualizing Key Signaling Pathways
The following diagrams illustrate the core signaling pathways of apoptosis and necroptosis, highlighting how chemotherapeutic agents can engage these molecular cascades.
The Scientist's Toolkit: Essential Research Reagents
The following table details key reagents and inhibitors used in the featured research to elucidate and manipulate cell death pathways.
Table 3: Key Reagents for Cell Death Research
| Reagent / Inhibitor | Primary Function / Target | Application in Cell Death Research |
|---|---|---|
| zVAD-FMK | Pan-caspase inhibitor [131] | Used to inhibit apoptosis and determine if cell death proceeds via alternative, caspase-independent pathways (e.g., necroptosis) [131] [127]. |
| Necrostatin-1 (Nec-1) | Specific inhibitor of RIPK1 [131] | Used to selectively inhibit the necroptosis pathway and evaluate its contribution to overall cell death [131]. |
| Ferrostatin-1 (Fer-1) | Potent ferroptosis inhibitor; acts as a radical scavenger [131] | Used to protect cells from lipid peroxidation and confirm the induction of ferroptosis [131] [130]. |
| Annexin V (FITC conjugate) | Binds to externalized phosphatidylserine (PS) [126] | A key marker for detecting early-stage apoptosis via flow cytometry or microscopy, often used in conjunction with PI [126]. |
| Propidium Iodide (PI) | DNA intercalating dye that is membrane impermeant | Used to label cells with compromised plasma membrane integrity, identifying late apoptotic/necrotic cells in conjunction with Annexin V [126]. |
| Antibodies against Cleaved Caspase-3 | Detects activated caspase-3 [131] | A standard immunohistochemical and western blot marker for confirming the execution phase of apoptosis [131]. |
| Antibodies against pMLKL | Detects phosphorylated MLKL [131] | A critical marker for confirming the activation of the necroptosis pathway in western blot analyses [131]. |
| Erastin | System Xc- inhibitor, induces ferroptosis [130] | Used as a positive control to induce ferroptosis or in combination studies to sensitize cells to ferroptotic death [130]. |
Establishing a Multi-Parameter Framework for Definitive Classification
Within cell biology and oncology drug screening, the precise differentiation between apoptosis and necrosis is not merely academic; it is a fundamental prerequisite for accurate therapeutic evaluation and mechanistic understanding [19]. While both are major forms of cell death, they are distinguished by unique morphological, biochemical, and functional characteristics, with apoptosis being a highly regulated, caspase-dependent process and necrosis traditionally viewed as an unregulated, accidental form of death [2] [134]. The challenge for researchers and drug development professionals lies in moving beyond single-parameter assays, which often fail to provide conclusive discrimination, particularly when cells undergo secondary necrosis [19]. This guide establishes a multi-parameter framework for definitive classification, objectively comparing the performance of traditional and advanced methodological alternatives to equip scientists with the tools for unambiguous cell death identification.
Comparative Analysis of Cell Death Features
A definitive classification system must be rooted in the core morphological and biochemical disparities that define each cell death type. The table below provides a consolidated comparison of these critical parameters.
Table 1: Key Characteristics of Apoptosis and Necrosis
| Parameter | Apoptosis | Necrosis |
|---|---|---|
| Morphology | Cell shrinkage, chromatin condensation (pyknosis), nuclear fragmentation (karyorrhexis), formation of apoptotic bodies, intact organelles [2] [3] [134]. | Cell and organelle swelling (oncosis), loss of plasma membrane integrity, disintegration of structures [2] [3] [135]. |
| Cell Membrane | Membrane blebbing with intact integrity; phosphatidylserine (PS) externalization [134]. | Loss of membrane integrity and increased permeability; eventual rupture [134] [135]. |
| Inflammatory Response | Typically no inflammation due to rapid phagocytosis of apoptotic bodies [2]. | Prominent inflammatory response triggered by release of intracellular contents [2] [3]. |
| Key Biochemical Hallmarks | Caspase activation (especially caspase-3), DNA laddering, cytochrome c release, energy (ATP)-dependent [19] [2] [134]. | Caspase-independent, random DNA degradation, ATP-independent [134] [135]. |
| Primary Inducers | Physiological developmental signals, mild cellular damage, chemotherapeutic agents [2]. | Extreme physical/chemical stress, toxins, ischemia, infection [134] [135]. |
Experimental Protocols for Definitive Classification
FRET-Based Live-Cell Imaging for Real-Time Discrimination
This sensitive live-cell method allows for the distinction between apoptosis and necrosis at a single-cell level in real time, making it a powerful tool for dynamic drug screening [19].
- Core Principle: The protocol uses cancer cells stably expressing two genetically encoded probes: a FRET-based caspase sensor (ECFP and EYFP linked by a DEVD caspase-cleavable sequence) and a mitochondrial-targeted DsRed protein. Caspase activation is detected by a loss of FRET, while necrosis is identified by the loss of the soluble FRET probe while retaining mitochondrial fluorescence [19].
- Detailed Methodology:
- Cell Line Development: Generate stable cell lines (e.g., neuroblastoma, U251) expressing the FRET-based caspase probe and Mito-DsRed. Select single-cell clones with homogenous expression for consistent imaging [19].
- Treatment and Imaging: Plate cells and treat with compounds of interest (e.g., 20 µM camptothecin and 50 µM etoposide for apoptosis; 5% peroxide bleach for necrosis). Perform real-time imaging using wide-field, confocal, or high-throughput microscopy platforms [19].
- Image Acquisition: Collect donor (ECFP) and acceptor (EYFP) fluorescence channels simultaneously with the Mito-DsRed channel at regular intervals (e.g., every 15-30 minutes) over 24-48 hours [19].
- Data Analysis and Classification:
- Viable Cell: Intact FRET probe (no ratio change) and retained Mito-DsRed fluorescence.
- Apoptotic Cell: Loss of FRET (increased ECFP/EYFP ratio) with retained Mito-DsRed fluorescence.
- Necrotic Cell: Loss of FRET probe fluorescence (no ratio change) with retained Mito-DsRed fluorescence [19].
Multimodal Optical Microscopy and Automated Classification
This approach leverages quantitative features from multiple co-registered imaging modalities to create an automated classification system [136].
- Core Principle: A support vector machine (SVM) classifier is trained on a 3D feature vector from each pixel—comprising Two-Photon Excitation Fluorescence (TPEF) intensity, Fluorescence Lifetime Imaging Microscopy (FLIM) of NADH, and Optical Coherence Microscopy (OCM) scattering intensity—to distinguish cell death modes [136].
- Detailed Methodology:
- Sample Preparation and Treatment: Use engineered tissue models (e.g., living skin). Induce apoptosis with a cocktail like 20 µM camptothecin and 50 µM etoposide; induce necrosis with 5% peroxide bleach [136].
- Multimodal Image Acquisition: Image samples with a custom microscope capable of simultaneous TPEF, FLIM, and OCM. For example, excite with a 730 nm titanium-sapphire laser and detect signals with a 16-channel PMT spectrometer for TPEF/FLIM and a spectrometer with a CCD camera for OCM [136].
- Image Processing:
- Segmentation: Use a tool like CellProfiler to segment cellular regions based on TPEF intensity images.
- Feature Extraction: At each pixel, construct a feature vector:
[OCM Intensity, TPEF Intensity, Mean Fluorescence Lifetime][136].
- Machine Learning Classification: Train an SVM model on a labeled dataset to find the optimal hyperplane separating apoptosis, necrosis, and viable cells in the 3D feature space. Validate the model's accuracy using 10-fold cross-validation [136].
Performance Data and Comparative Efficacy
The following table summarizes quantitative data and performance metrics for the key methodologies discussed, providing an objective comparison for researchers.
Table 2: Experimental Data and Performance Comparison of Classification Methods
| Methodology | Key Measurable Parameters | Reported Outcome / Performance | Advantages | Limitations |
|---|---|---|---|---|
| FRET-Based Live-Cell Imaging [19] | - FRET ratio (ECFP/EYFP)- Mito-DsRed fluorescence retention- Temporal dynamics of caspase activation | Discriminates primary necrosis, apoptosis, and secondary necrosis at single-cell resolution. Adaptable to high-throughput screening. | Real-time, single-cell kinetic data; visual confirmation of morphology; highly sensitive. | Requires generation of stable cell lines; potential for phototoxicity during long-term imaging. |
| Multimodal Microscopy + SVM [136] | - TPEF intensity (NADH)- FLIM lifetime- OCM scattering intensity | High classification accuracy (>90% reported) for apoptotic vs. necrotic cells in engineered skin tissue using 10-fold cross-validation. | Label-free, quantitative; provides metabolic context (via NADH); automated, unbiased classification. | Requires specialized, expensive equipment; complex data processing pipeline. |
| Annexin V/PI Staining [19] [134] | - Phosphatidylserine exposure (Annexin V)- Membrane integrity (Propidium Iodide) | Widely used but can misclassify late apoptotic/secondary necrotic cells as primary necrotic; snapshot in time. | Simple, accessible, low-cost. | Inability to provide real-time kinetics; difficult to distinguish apoptosis from secondary necrosis. |
Signaling Pathways and Experimental Workflow
The molecular pathways governing apoptosis and necrosis are distinct. The following diagrams, generated using the specified color palette, illustrate these pathways and a generalized experimental workflow.
Apoptotic and Necroptotic Signaling Pathways
Diagram 1: Apoptosis and Necroptosis Pathways
Multimodal Imaging Classification Workflow
Diagram 2: Multimodal Imaging and Analysis Workflow
The Scientist's Toolkit: Research Reagent Solutions
The following table details essential reagents and tools for implementing the described multi-parameter frameworks.
Table 3: Essential Research Reagents and Tools for Cell Death Classification
| Item | Function / Application | Specific Example / Target |
|---|---|---|
| FRET-Based Caspase Probe [19] | Genetically encoded sensor for real-time detection of caspase activation. Cleavage leads to loss of FRET. | ECFP-DEVD-EYFP construct. |
| Organelle-Targeted Fluorescent Protein [19] | Serves as a cell viability and morphology marker; retained in necrotic cells after loss of cytosolic probes. | Mitochondrially-targeted DsRed (Mito-DsRed). |
| Caspase Antibodies [134] | Immunodetection of caspase activation, a hallmark of apoptosis, via techniques like IHC and WB. | Anti-Caspase-3 (cleaved), Anti-BAX. |
| BAX/BAK Antibodies [134] | Detect activation of pro-apoptotic BCL-2 family proteins, key regulators of intrinsic apoptosis. | Anti-BAX, Anti-BAK. |
| Annexin V Conjugates [134] | Binds to phosphatidylserine (PS) exposed on the outer leaflet of the plasma membrane in early apoptosis. | FITC- or PE-conjugated Annexin V. |
| Viability Dyes [134] [135] | Assess plasma membrane integrity. Impermeant to live and early apoptotic cells; stain necrotic cells. | Propidium Iodide (PI), 7-AAD. |
| Chemical Inducers [19] [136] | Positive controls for inducing specific cell death pathways in experimental models. | Camptothecin/Etoposide (Apoptosis), H₂O₂ (Necrosis). |
| MLKL & RIP3 Antibodies [134] | Key biomarkers for the detection of necroptosis, a regulated form of necrotic cell death. | Anti-MLKL, Anti-RIP3. |
Conclusion
The precise discrimination between apoptosis and necrosis based on morphological features remains a cornerstone of pathological assessment and experimental biology. While classical hallmarks provide a reliable foundation, the advent of advanced imaging and analytical techniques allows for a more dynamic and nuanced understanding. The growing complexity of the cell death landscape, particularly with the characterization of regulated forms like necroptosis, necessitates a multi-parametric approach that integrates morphology with biochemical and molecular data. For researchers and drug developers, mastering this discrimination is not merely academic; it is critical for accurately evaluating the mechanisms and efficacy of therapeutic interventions, especially in oncology. Future directions will likely involve the development of more sophisticated, automated imaging analysis tools and a deeper exploration of how these distinct cell death modalities shape the tumor microenvironment and overall treatment outcomes.
References
- 1. Necrosis Vs. Apoptosis: Processes, Necrotic Cell Death ... [https://www.akadeum.com/blog/necrosis-vs-apoptosis-processes-necrotic-cell-death-apoptosis-steps/?srsltid=AfmBOooGjJOF-x0QqSgr7GSrqMB0hhDfWEjTBAW5JcXiAzEHdzGsXNKK]
- 2. Apoptosis: A Review of Programmed Cell Death - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2117903/]
- 3. Research progress on morphology and mechanism of ... [https://www.nature.com/articles/s41419-024-06712-8]
- 4. 50 years on and still very much alive: 'Apoptosis [https://www.nature.com/articles/s41416-022-02020-0]
- 5. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOorKf-fnBmdnglxS7wtXiJJpiWWxBQ36DNuFSYnz_bkxrGuiAYe7]
- 6. Morphological assessment of apoptosis [https://www.sciencedirect.com/science/article/abs/pii/S1046202307002150]
- 7. High-Resolution Imaging of Morphological Changes ... [https://www.mdpi.com/2079-6374/15/8/522]
- 8. Uncoupling Cell Shrinkage from Apoptosis Reveals That ... [https://www.sciencedirect.com/science/article/pii/S0021925820832359]
- 9. Necrosis Vs. Apoptosis: Processes, Necrotic Cell Death ... [https://www.akadeum.com/blog/necrosis-vs-apoptosis-processes-necrotic-cell-death-apoptosis-steps/?srsltid=AfmBOoq6ADoH9K8oXQ3X021O9S6bExMcUD_2Ln40xKonqknkuCUjHO1c]
- 10. Diversity and complexity of cell death: a historical review [https://www.nature.com/articles/s12276-023-01078-x]
- 11. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOopWTjzjqWJ3NvJEaB1a6iIjm-ZWsZ1KfkVBGF_0unHzsvZnGEaW]
- 12. ApoNecV: A macro for cell death type differentiation - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11891952/]
- 13. Identification of the pyroptosis, apoptosis, and necroptosis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12211273/]
- 14. Pyknosis [https://en.wikipedia.org/wiki/Pyknosis]
- 15. Karyolysis - an overview [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/karyolysis]
- 16. Necrosis Vs. Apoptosis: Processes, Necrotic Cell Death ... [https://www.akadeum.com/blog/necrosis-vs-apoptosis-processes-necrotic-cell-death-apoptosis-steps/?srsltid=AfmBOoqdATTUcrFfKtehhvbdde1AnvzH0EPYh2j6CwM661s4r1zp8bPn]
- 17. BIOMARKERS DISTINGUISH APOPTOTIC AND NECROTIC ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4213307/]
- 18. A Comparative Study of Apoptosis and Necrosis in HepG2 ... [https://www.sciencedirect.com/science/article/pii/S0006291X98901395]
- 19. A quantitative real-time approach for discriminating ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5253725/]
- 20. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOopfP0V0L1uK-D3oOqBBf37G-1hvgybyZ3L8jO96TtCrj38p2ko3]
- 21. Comprehensive landscape of cell death mechanisms [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1611055/full]
- 22. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOooOrSiXpq_TDUL0v5NlBCmfomtJwFb_Yb2Q7KPlyCY25h1cBvhi]
- 23. Necrosis Vs. Apoptosis: Processes, Necrotic Cell Death ... [https://www.akadeum.com/blog/necrosis-vs-apoptosis-processes-necrotic-cell-death-apoptosis-steps/?srsltid=AfmBOoq9HBdHGazyfLa-Nl9Gl2RA51AjlbTD0CsxbktgN92Zb1mhda6H]
- 24. Plasma membrane changes during programmed cell deaths [https://www.nature.com/articles/cr2017133]
- 25. Necrosis: a specific form of programmed cell death? [https://pubmed.ncbi.nlm.nih.gov/12565815/]
- 26. Plasma membrane permeabilization following cell death [https://pmc.ncbi.nlm.nih.gov/articles/PMC8289853/]
- 27. Relationship between membrane permeability and ... [https://www.sciencedirect.com/science/article/pii/S0005273611001064]
- 28. Plasma membrane perforation by GSDME during ... [https://link.springer.com/article/10.1007/s00018-021-04078-0]
- 29. Cell death signaling and immune regulation - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC12355773/]
- 30. Apoptosis - StatPearls - NCBI Bookshelf [https://www.ncbi.nlm.nih.gov/sites/books/NBK499821/]
- 31. Apoptosis: A Comprehensive Overview of Signaling ... [https://www.mdpi.com/2073-4409/13/22/1838]
- 32. Necrosis Pathology - StatPearls - NCBI Bookshelf - NIH [https://www.ncbi.nlm.nih.gov/books/NBK557627/]
- 33. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOoot0ujdL98gGrV2mlFrJzTBZHXRQLQRshfmpCsAq60Kw7Zm2xG7]
- 34. Necroptosis, pyroptosis and apoptosis: an intricate game ... [https://www.nature.com/articles/s41423-020-00630-3]
- 35. A quantitative real-time approach for discriminating ... [https://www.nature.com/articles/cddiscovery2016101]
- 36. Comparison of necrotizing enterocolitis (NEC) [https://pmc.ncbi.nlm.nih.gov/articles/PMC12267699/]
- 37. Regulated necrosis: disease relevance and therapeutic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6531857/]
- 38. Necrosis Vs. Apoptosis: Processes, Necrotic Cell Death ... [https://www.akadeum.com/blog/necrosis-vs-apoptosis-processes-necrotic-cell-death-apoptosis-steps/?srsltid=AfmBOopOLHIYQ8UUTey6ArHz5dLoi_FUmS5jn2tq-t18UYOdsVBYx9Hl]
- 39. Regulated necrosis, a proinflammatory cell death ... [https://www.nature.com/articles/s41419-022-05066-3]
- 40. Exploring necroptosis: mechanistic analysis and antitumor ... [https://www.nature.com/articles/s41420-025-02423-x]
- 41. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOor3qcXO5IT_BAQBkutOhwg6IyslqCBJqsImmUiEX9uLyybxISM5]
- 42. Identification and experimental verification of necroptosis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12579482/]
- 43. Necroptosis: The Release of Damage-Associated ... [https://www.sciencedirect.com/science/article/pii/S1074761313000587]
- 44. Restoring apoptosis in breast cancer via peptide mediated ... [https://www.nature.com/articles/s41598-025-24772-4]
- 45. High-Resolution Imaging of Morphological Changes ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12384366/]
- 46. Dynamic contrast optical coherence tomography (DyC ... [https://www.nature.com/articles/s42003-024-05973-5]
- 47. Dynamic full-field optical coherence tomography module ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10539404/]
- 48. Label-free cellular imaging of mouse retina with dual-mode ... [https://www.opticsjournal.net/Articles/OJ40ed3773dcc1eee9/FullText]
- 49. optical transmission tomography for biosystems... Label free [https://pmc.ncbi.nlm.nih.gov/articles/PMC9408247/]
- 50. Continuous scanning full-field OCT for fast volumetric imaging of... [https://arxiv.org/html/2509.08101v1]
- 51. High-Resolution Imaging of Morphological Changes ... [https://pubmed.ncbi.nlm.nih.gov/40862983/]
- 52. Apoptosis vs Necrosis [http://www.cyto.purdue.edu/sites/default/files/__subsites__/archive/flowcyt/research/cytotech/apopto/data/chap10.htm]
- 53. Label-Free Three-Dimensional Morphological ... [https://www.mdpi.com/1424-8220/24/11/3435]
- 54. Deep learning with digital holographic microscopy ... [https://www.nature.com/articles/s41420-021-00616-8]
- 55. Quantitative analysis and comparison of 3D morphology ... [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0184726]
- 56. Comparison of Several Techniques for the Detection ... [https://www.creative-bioarray.com/support/comparison-of-several-techniques-for-the-detection-of-apoptotic-cells.htm]
- 57. Characterizing Cell Adhesion by Using Micropipette Aspiration [https://pmc.ncbi.nlm.nih.gov/articles/PMC4621874/]
- 58. How Cells Tiptoe on Adhesive Surfaces before Sticking [https://www.sciencedirect.com/science/article/pii/S0006349508704119]
- 59. Comparative Analysis of Different Methodological ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1867030/]
- 60. Detection of drug-induced apoptosis and necrosis in ... [https://www.nature.com/articles/4400802]
- 61. Detection of apoptosis and necrosis in normal human lung ... [https://hub.tmu.edu.tw/en/publications/detection-of-apoptosis-and-necrosis-in-normal-human-lung-cells-us/]
- 62. Metabolic fingerprinting by nuclear magnetic resonance of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12356598/]
- 63. Apoptosis, Pyroptosis, and Necrosis: Mechanistic Description ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC1087413/]
- 64. Caspases as master regulators of programmed cell death [https://www.nature.com/articles/s12276-025-01470-9]
- 65. Programmed Cell-Death Mechanism Analysis Using Same ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8679084/]
- 66. Helminthic larval stage induces cellular apoptosis via caspase ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12507911/]
- 67. Extrinsic apoptosis and necroptosis in telencephalic ... [https://www.nature.com/articles/s41418-025-01594-5]
- 68. Programmed cell death detection methods: a systematic review and a categorical comparison | Apoptosis [https://link.springer.com/article/10.1007/s10495-022-01735-y]
- 69. Apoptosis Marker Assays for HTS - NCBI [https://www.ncbi.nlm.nih.gov/books/NBK572437/]
- 70. Morphologic and biochemical hallmarks of apoptosis [https://pubmed.ncbi.nlm.nih.gov/10728374/]
- 71. Apoptosis/ Necrosis Assay Kit (blue, green, red) (ab176749) | Abcam [https://www.abcam.com/en-us/products/assay-kits/apoptosis-necrosis-assay-kit-blue-green-red-ab176749]
- 72. Necrosis vs Apoptosis Assay Kit | 9148 [https://www.antibodiesinc.com/products/necrosis-vs-apoptosis-assay-kit-9147]
- 73. Secondary necrosis in multicellular animals: an outcome of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7102248/]
- 74. Secondary necrosis: The natural outcome of the complete ... [https://www.sciencedirect.com/science/article/pii/S0014579310008641]
- 75. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOoooOdVS8icALgU_tDf2mOqVPbBIaL7Hj4c7hm8oCe4FrLYtOLG5]
- 76. Necrosis Vs. Apoptosis: Processes, Necrotic Cell Death ... [https://www.akadeum.com/blog/necrosis-vs-apoptosis-processes-necrotic-cell-death-apoptosis-steps/?srsltid=AfmBOor3mDA1_vFvOQ6Hl_dJ1U_WbNksmDHD1w7P6smMu5t9AZjacq_C]
- 77. Apoptosis vs Necrosis - Difference and Comparison [https://www.diffen.com/difference/Apoptosis_vs_Necrosis]
- 78. Necroptosis, necrosis and secondary necrosis converge on ... [https://www.nature.com/articles/cdd2009184]
- 79. Imaging of morphological and biochemical hallmarks ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6851291/]
- 80. Researchers identify new molecular switch involved in ... [https://www.news-medical.net/news/20251103/Researchers-identify-new-molecular-switch-involved-in-programmed-cell-death.aspx]
- 81. Mechanisms of Cell Death: Necrosis & Necroptosis [https://blog.cellsignal.com/mechanisms-of-cell-death-necrosis-necroptosis]
- 82. Necrosis vs Necroptosis vs Apoptosis [https://www.antibodiesinc.com/pages/necrosis-vs-necroptosis-vs-apoptosis?srsltid=AfmBOooPUT5EJmDF6NJ2i--Tom-733cDgYl1EmaMHoMXR91Sx9m2mUXR]
- 83. Pyroptosis versus necroptosis: similarities, differences, and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6294779/]
- 84. Apoptosis vs Necroptosis: Identifying Both Types of Cell ... [https://www.bio-techne.com/resources/blogs/apoptosis-and-necroptosis-part-i-important-factors-to-identify-both-types-of-programmed-cell-death]
- 85. Discriminating Between Apoptosis, Necrosis, Necroptosis ... [https://pubmed.ncbi.nlm.nih.gov/38112058/]
- 86. Caspase inhibitors promote alternative cell death pathways [https://pubmed.ncbi.nlm.nih.gov/17062895/]
- 87. A long way to go: caspase inhibitors in clinical use [https://www.nature.com/articles/s41419-021-04240-3]
- 88. Alternative cell death mechanisms in development and beyond [https://genesdev.cshlp.org/content/24/23/2592.full.html]
- 89. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOop6U1_nx3R2yYM_4u5RFm--L02YXPnBG5hd068J3GpvHqF12CLe]
- 90. Necrosis Vs. Apoptosis: Processes, Necrotic Cell Death ... [https://www.akadeum.com/blog/necrosis-vs-apoptosis-processes-necrotic-cell-death-apoptosis-steps/?srsltid=AfmBOor1M_g6OVlx73qkV1H1UxLg-qSqRi7dRFaZE43kG6usdUUVvgID]
- 91. Frontiers | Cell death in acute lung injury: caspase -regulated... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1559659/full]
- 92. Caspases as Targets for Drug Development - NCBI - NIH [https://www.ncbi.nlm.nih.gov/books/NBK6578/]
- 93. Activation and Caspase -mediated Inhibition of PARP: A Molecular... [https://pmc.ncbi.nlm.nih.gov/articles/PMC99613/]
- 94. Cell death in mammalian cell culture: molecular mechanisms and cell line engineering strategies - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2932912/]
- 95. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOopnihgFY_wCZGL-g8VsDS2Pzm4btf6L7lo08Jme1B84jQ5M7S3a]
- 96. Different types of cell death and their shift in shaping disease [https://www.nature.com/articles/s41420-023-01581-0]
- 97. Morphological criteria to distinguish cell death induced by apoptotic and necrotic treatments | Apoptosis [https://link.springer.com/article/10.1007/s10495-005-6075-6]
- 98. Evaluating cell viability assessment techniques: a comparative ... [https://biomedical-engineering-online.biomedcentral.com/articles/10.1186/s12938-025-01452-y]
- 99. Integrated real-time imaging of executioner caspase ... [https://www.nature.com/articles/s41420-025-02662-y]
- 100. Molecular Imaging of Apoptosis: The Case of Caspase-3 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8069194/]
- 101. A comparison of cell death pathways in three different ... [https://pubmed.ncbi.nlm.nih.gov/40751929/]
- 102. Identification of the pyroptosis, apoptosis, and necroptosis ... [https://eurjmedres.biomedcentral.com/articles/10.1186/s40001-025-02702-4]
- 103. Endoplasmic reticulum–mitochondria contacts are prime ... [https://www.nature.com/articles/s41556-025-01668-z]
- 104. Investigating apoptosis in peripheral blood mononuclear cells ... [https://bmcimmunol.biomedcentral.com/articles/10.1186/s12865-025-00769-6]
- 105. Learning and actioning general principles of cancer cell ... [https://www.nature.com/articles/s41467-025-56827-5]
- 106. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOoofordZnzGdAe8cR2l9trR1FDWTBBdTmJaRHa8bblSeUQb5bGDR]
- 107. Necrosis vs Necroptosis vs Apoptosis [https://www.antibodiesinc.com/pages/necrosis-vs-necroptosis-vs-apoptosis?srsltid=AfmBOopDG9ukuzDQbGcUQCOGfhalISYFqD3SLcSXA5DJzaqlhdK-09fU]
- 108. RIPK1 in necroptosis and recent progress in related ... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1480027/full]
- 109. Regulated cell death pathways and their roles in ... - Nature [https://www.nature.com/articles/s12276-023-01069-y]
- 110. Targeting regulated cell death: Apoptosis, necroptosis ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11921819/]
- 111. Necrosis Vs. Apoptosis: Processes, Necrotic Cell Death ... [https://www.akadeum.com/blog/necrosis-vs-apoptosis-processes-necrotic-cell-death-apoptosis-steps/?srsltid=AfmBOopU4zl0eUngXCaxWdeZypzDu4HzMI5vP2tB12BR5Q2F9wLPFIqi]
- 112. RIPK1 in necroptosis and recent progress in related ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11850271/]
- 113. Apoptosis: A Comprehensive Overview of Signaling Pathways ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11592877/]
- 114. RIPK1 signaling pathways: implications for autoimmune and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12466156/]
- 115. FDA-approved phensuximide inhibits RIPK1-dependent ... [https://www.nature.com/articles/s41419-025-07754-2]
- 116. Pharmacological and Mechanism Study of a New RIPK1 ... [https://www.sciencedirect.com/science/article/abs/pii/S0006497124035572]
- 117. A Comparative Analysis of Pediatric and Adult ICU Patients [https://www.mdpi.com/2227-9059/13/7/1747]
- 118. Apoptosis Detection [https://www.bdbiosciences.com/en-us/learn/research/cell-biology/apoptosis]
- 119. Non-invasive detection and differentiation of apoptotic and ... [https://www.sciencedirect.com/science/article/pii/S1011134423000842]
- 120. LANCE: a Label-Free Live Apoptotic and Necrotic Cell ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10729583/]
- 121. The selection between apoptosis and necrosis is ... [https://www.nature.com/articles/4401249]
- 122. Flow cytometry-based apoptosis detection - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3863590/]
- 123. A comparison of cell death pathways in three different kinds of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12320871/]
- 124. Quantification of apoptosis and necroptosis at the single ... [https://www.sciencedirect.com/science/article/abs/pii/S0022175915001520]
- 125. Insights on the crosstalk among different cell death ... [https://www.nature.com/articles/s41420-025-02328-9]
- 126. DNA-protein cross-links emerge as major contributors to ... [https://www.nature.com/articles/s41598-025-06705-3]
- 127. Apoptosis, necroptosis, and pyroptosis as alternative cell ... [https://www.sciencedirect.com/science/article/pii/S0304419X23001737]
- 128. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOorVCPV3ko7AZU0eGLAYJVSfe9wdyQicU8HE69iwdf0vs8XDAmVF]
- 129. Necrosis Vs. Apoptosis: Processes, Necrotic Cell Death ... [https://www.akadeum.com/blog/necrosis-vs-apoptosis-processes-necrotic-cell-death-apoptosis-steps/?srsltid=AfmBOooYyrPpGvqzwrgLO0YVwKE2SB1OCzBNOojVbwIZKywxq6KNWxmH]
- 130. Research progress on ferroptosis and PARP inhibitors in ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1598279/full]
- 131. Pharmacological inhibition of apoptosis, necroptosis, and ... [https://www.nature.com/articles/s41598-025-17275-9]
- 132. Recent Tumor Necrosis Factor-Related Apoptosis-Inducing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11922527/]
- 133. Drug discovery for chemotherapeutic resistance based on ... [https://www.frontiersin.org/journals/bioinformatics/articles/10.3389/fbinf.2025.1661601/full]
- 134. What is the difference between necrosis and apoptosis? [https://www.ptglab.com/news/blog/what-is-the-difference-between-necrosis-and-apoptosis/?srsltid=AfmBOoq3sxVDvhWHkZi6nk2otoKW2NxxORFrFBflX1mFHkWYjZCcUIhY]
- 135. Necrosis Vs. Apoptosis: Processes, Necrotic Cell Death ... [https://www.akadeum.com/blog/necrosis-vs-apoptosis-processes-necrotic-cell-death-apoptosis-steps/?srsltid=AfmBOopNO6pNyQygLR5M3nPzrEi22RD_4GbPd8u55AEJeVpUO_C1AAVC]
- 136. A quantitative framework for the analysis of multimodal optical ... [https://qims.amegroups.org/article/view/13783/html]