Monitoring Apoptosis in Real-Time: The GFP-Cytochrome c Reporter System Explained
This article provides a comprehensive guide for researchers on using GFP (Green Fluorescent Protein) reporter systems to detect cytochrome c localization during apoptosis.
Monitoring Apoptosis in Real-Time: The GFP-Cytochrome c Reporter System Explained
Abstract
This article provides a comprehensive guide for researchers on using GFP (Green Fluorescent Protein) reporter systems to detect cytochrome c localization during apoptosis. We cover the foundational biology of cytochrome c release, detailed methodologies for constructing and using GFP-cyt c fusions, common troubleshooting and optimization techniques for live-cell imaging, and validation strategies comparing this approach to other assays. Aimed at scientists and drug development professionals, this resource integrates current protocols and best practices for applying this critical tool in cell death research and therapeutic screening.
Cytochrome c and Apoptosis: Why Tracking Its Release is Fundamental to Cell Death Research
The Pivotal Role of Cytochrome c in the Intrinsic Apoptotic Pathway
Within the broader context of developing a GFP reporter system for detecting cytochrome c (Cyt c) subcellular localization, understanding its precise role in apoptosis is foundational. This whitepaper provides a technical dissection of Cyt c's function in the intrinsic apoptotic pathway, serving as a critical reference for researchers utilizing localization assays in mechanistic studies and drug discovery.
Biochemical Release and Activation Cascade
Cyt c is a nuclear-encoded mitochondrial hemoprotein. Upon integration of diverse apoptotic stimuli (e.g., DNA damage, oxidative stress), mitochondrial outer membrane permeabilization (MOMP) occurs, primarily regulated by Bcl-2 family proteins. Cyt c is released from the mitochondrial intermembrane space into the cytosol.
Its pivotal role is initiated upon cytosolic binding to Apoptotic Protease Activating Factor 1 (Apaf-1). This binding, in the presence of dATP/ATP, induces a conformational change in Apaf-1, triggering its oligomerization into a wheel-like complex known as the apoptosome.
Table 1: Key Quantitative Parameters of Cytochrome c and Apoptosome Formation
| Parameter | Value / Measurement | Experimental Context / Notes |
|---|---|---|
| Molecular Weight of Cyt c | ~12.4 kDa | Horse heart Cyt c often used in in vitro studies. |
| Concentration for Apoptosome Assembly in vitro | 0.1 - 10 µM | Varies with Apaf-1 and nucleotide concentration. |
| Optimal Nucleotide | dATP > ATP | dATP (10-100 µM) is more potent in supporting apoptosome assembly. |
| Apaf-1 Oligomer Stoichiometry | Heptamer | Forms a heptameric complex upon Cyt c/dATP binding. |
| Caspase-9 Activation Kd | Low nanomolar range | For binding to the apoptosome platform. |
Diagram 1: Cytochrome c-Mediated Apoptosome Formation Pathway
Experimental Protocols for Key Assays
In VitroApoptosome Reconstitution & Caspase Activation Assay
Purpose: To demonstrate the essential role of Cyt c in activating caspase-9 via apoptosome assembly. Materials: Purified recombinant human Apaf-1, horse heart cytochrome c, dATP, recombinant procaspase-9, colorimetric caspase-9 substrate (e.g., Ac-LEHD-pNA). Protocol:
- Prepare reaction buffer: 20 mM HEPES-KOH (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.1% CHAPS.
- In a 96-well plate, mix Apaf-1 (50 nM), cytochrome c (1 µM), and dATP (1 mM) in reaction buffer. Incubate at 30°C for 60 min to allow apoptosome assembly.
- Add procaspase-9 (20 nM) to the mixture and incubate at 30°C for an additional 30 min.
- Initiate the catalytic reaction by adding the caspase-9 substrate Ac-LEHD-pNA (final 200 µM).
- Monitor the absorbance at 405 nm continuously for 60-120 min using a plate reader. The rate of p-nitroaniline (pNA) release is proportional to caspase-9 activity.
- Controls: Omit cytochrome c or dATP in negative control reactions.
Immunofluorescence Protocol for Cytochrome c Release Detection (GFP Reporter Context)
Purpose: To visualize the translocation of cytochrome c from mitochondria to cytosol in fixed cells, often validated against a GFP-tagged Cyt c construct. Materials: Cells grown on coverslips, anti-cytochrome c antibody (clone 6H2.B4), fluorescent secondary antibody (e.g., Alexa Fluor 594), MitoTracker Deep Red, paraformaldehyde (4%), Triton X-100, blocking serum. Protocol:
- Induction & Staining: Treat cells with apoptosis inducer (e.g., 1 µM Staurosporine, 4-6 hrs). Prior to fixation, incubate with MitoTracker Deep Red (50 nM) in culture medium for 30 min at 37°C.
- Fixation & Permeabilization: Wash cells with PBS and fix with 4% PFA for 15 min at RT. Wash, then permeabilize with 0.1% Triton X-100 in PBS for 10 min.
- Blocking & Antibody Incubation: Block with 3% BSA in PBS for 1 hr. Incubate with primary anti-cytochrome c antibody (1:200 in blocking buffer) overnight at 4°C.
- Visualization: Wash and incubate with Alexa Fluor 594-conjugated secondary antibody (1:500) for 1 hr at RT in the dark. Wash thoroughly.
- Mounting & Imaging: Mount coverslips using anti-fade mounting medium with DAPI. Image using a confocal microscope.
- Interpretation: Non-apoptotic cells show punctate mitochondrial staining (co-localized with MitoTracker). Apoptotic cells show a diffuse cytosolic fluorescence pattern for Cyt c, distinct from mitochondrial marker.
Table 2: Research Reagent Solutions for Cytochrome c Localization Studies
| Reagent / Material | Function / Purpose | Example / Note |
|---|---|---|
| GFP-Cytochrome c Plasmid | Live-cell reporter for visualizing Cyt c localization dynamics in real-time. | Clone Cyt c cDNA into pEGFP-N1 vector. Mutations (e.g., K72A) can be used to study import/export. |
| Anti-Cytochrome c Antibody (6H2.B4) | Immunodetection of endogenous Cyt c release in fixed cells via IF/IHC. | Mouse monoclonal; works well for IF, WB, and IP. |
| MitoTracker Dyes (e.g., Deep Red) | Specific labeling of active mitochondria for co-localization reference. | Cell-permeant, fixes with aldehyde. Use before PFA fixation. |
| Apoptosis Inducers (Staurosporine, ABT-263) | Positive control stimuli to trigger intrinsic pathway and Cyt c release. | Staurosporine is a broad kinase inhibitor; ABT-263 (Navitoclax) is a BH3 mimetic. |
| Caspase-9 Colorimetric Assay Kit | Quantitative measurement of downstream apoptosome activity. | Contains Ac-LEHD-pNA substrate and assay buffer. |
| Recombinant Human Apaf-1 Protein | For in vitro reconstitution of the apoptosome. | Essential for mechanistic biochemical studies. |
| Cell Permeabilization Reagent (e.g., Digitonin) | Selective plasma membrane permeabilization for studying Cyt c release in situ. | Low concentration (e.g., 0.01%) releases cytosolic but not mitochondrial proteins. |
| Z-VAD-FMK (pan-Caspase Inhibitor) | Negative control to confirm caspase-dependent apoptotic events downstream of Cyt c release. | Irreversible inhibitor; pre-treatment blocks apoptotic morphology. |
Downstream Consequences and Therapeutic Implications
The apoptosome-bound caspase-9 cleaves and activates effector caspases-3 and -7, executing the terminal phase of apoptosis. Dysregulation of Cyt c release is implicated in cancer (insufficient apoptosis) and neurodegenerative diseases (excessive apoptosis).
Table 3: Disease Associations and Drug Targeting Related to Cytochrome c Release
| Disease Area | Dysregulation | Potential Therapeutic Target / Strategy |
|---|---|---|
| Cancer | Inhibited Cyt c release due to Bcl-2/Bcl-xL overexpression. | BH3 mimetics (Venetoclax, Navitoclax) promote MOMP and Cyt c release. |
| Neurodegeneration (e.g., ALS, AD) | Excessive Cyt c release contributes to neuronal loss. | Caspase inhibitors, MOMP inhibitors (e.g., targeting Bax/Bak). |
| Myocardial Infarction | Ischemia/reperfusion injury induces Cyt c release. | Cyclosporine A (inhibits mPTP opening upstream of MOMP). |
Diagram 2: Downstream Apoptotic Execution Pathway
GFP Reporter Applications and Validation
A GFP-Cyt c fusion construct is a vital tool for live-cell imaging of apoptosis. Its utility in high-content screening for pro- or anti-apoptotic compounds is significant. Key validation experiments include:
- Co-localization: Demonstrating GFP-Cyt c overlap with mitochondrial markers (e.g., MitoTracker) in healthy cells.
- Translocation Kinetics: Quantifying the time from stimulus to diffuse cytosolic signal using time-lapse microscopy.
- Correlation with Endogenous Event: Performing immunofluorescence on fixed, parallel samples with anti-Cyt c antibody to confirm GFP signal accurately reports endogenous protein localization.
This detailed mechanistic understanding, supported by robust experimental protocols, enables precise use of Cyt c localization as a definitive biomarker for intrinsic apoptotic engagement in basic research and drug development.
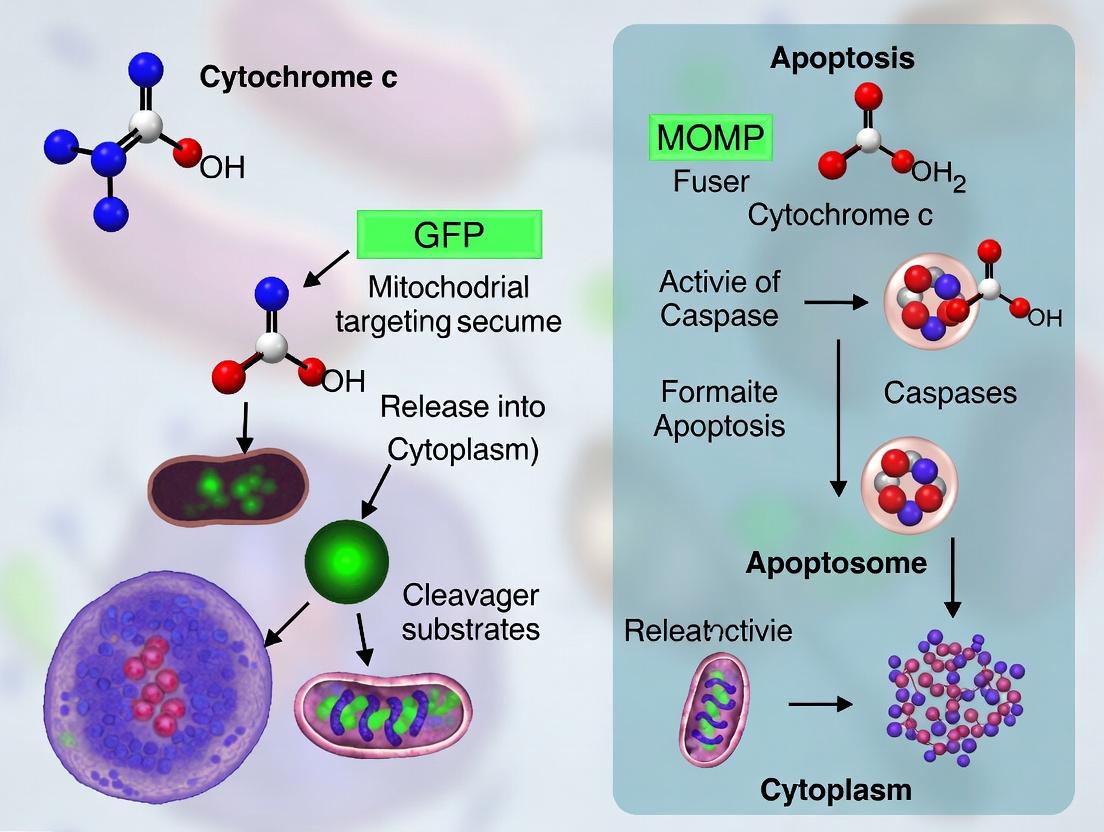
This whitepaper details the pivotal event in the intrinsic apoptosis pathway—the mitochondrial outer membrane permeabilization (MOMP) and subsequent cytochrome c release from the mitochondrial intermembrane space (IMS) into the cytosol. Framed within the context of developing and utilizing GFP-based reporters for visualizing this relocalization in real-time, this guide serves as a technical resource for researchers and drug development professionals aiming to quantify apoptotic commitment.
The Role of CytochromecRelocalization in Apoptotic Signaling
Cytochrome c, a component of the mitochondrial electron transport chain, is confined to the IMS in healthy cells. Upon apoptotic stimuli (e.g., DNA damage, oxidative stress), pro-apoptotic BCL-2 family proteins (e.g., BAX, BAK) oligomerize and permeabilize the MOM. The rupture of the MOM allows cytochrome c and other IMS proteins (e.g., SMAC/DIABLO) to diffuse into the cytosol. Cytosolic cytochrome c binds to APAF-1 and procaspase-9, forming the apoptosome, which activates caspase-9 and initiates the caspase cascade, leading to irreversible cell death.
GFP Reporters for Visualizing CytochromecRelease
The development of fluorescent protein reporters has been instrumental in visualizing this critical event in living cells. The core strategy involves tagging cytochrome c with a fluorescent protein (e.g., GFP) and monitoring its redistribution via live-cell microscopy.
Key Construct Designs
- Cytochrome c-GFP Fusion: Full-length cytochrome c is fused to the N- or C-terminus of GFP. The fusion protein is imported into mitochondria via cytochrome c's native mitochondrial targeting sequence and is functional in respiration.
- Split-Fluorescent Protein Systems: Cytochrome c is fused to a fragment of a split GFP (or Venus), while the complementary fragment is targeted to the IMS or cytosol. Reconstitution and fluorescence occur only upon cytochrome c release, increasing signal-to-noise.
- Biosensors with FRET Pairs: Cytochrome c is labeled with a FRET donor (e.g., CFP), and an MOM-anchored protein is labeled with a FRET acceptor (e.g., YFP). FRET loss indicates cytochrome c dissociation from the mitochondrion.
Experimental Considerations for GFP-CytochromecStudies
- Validation: The fusion protein must be validated to ensure it does not aberrantly induce apoptosis and is correctly localized and functional.
- Imaging Modalities: Confocal or widefield fluorescence microscopy with environmental control (CO₂, temperature) for time-lapse imaging.
- Quantification: Fluorescence intensity in cytosolic vs. mitochondrial regions over time is quantified to derive kinetic parameters of release.
Quantitative Data on CytochromecRelease Kinetics
Recent studies utilizing GFP-cytochrome c reporters have provided precise kinetic data on release events.
Table 1: Kinetic Parameters of Cytochrome c Release Following Various Apoptotic Stimuli
| Apoptotic Stimulus | Cell Line | Time to Initial Release (Mean ± SD) | Duration of Complete Release | % Cells Exhibiting "Full" Release | Key Measurement Method | Reference (Example) |
|---|---|---|---|---|---|---|
| Staurosporine (1 µM) | HeLa | 142 ± 28 min | ~30 min | >85% | Live-cell confocal, Cyto c-GFP | Goldstein et al., 2005 |
| UV-C Irradiation (50 J/m²) | MCF-7 | 285 ± 67 min | ~90 min | ~70% | Spinning-disk confocal, split-Venus | Waterhouse et al., 2014 |
| ABT-737 (1 µM) + S63845 (1 µM) | MV4;11 | 45 ± 15 min | <20 min | >95% | TIRF/Confocal, Cyto c-mCherry | Riley et al., 2021 |
| Tumor Necrosis Factor-α (w/ CHX) | HT-29 | 210 ± 45 min | ~60 min | ~80% | Widefield time-lapse, FRET-based sensor | Rehm et al., 2006 |
Table 2: Key Modulators of Cytochrome c Release and Their Effects
| Modulator/Target | Type | Effect on Cytochrome c Release Time/Cascade | Potential Therapeutic Context |
|---|---|---|---|
| Z-VAD-FMK | Pan-caspase inhibitor | Blocks downstream execution but does not prevent cytochrome c release | Tool compound to dissociate release from late apoptosis |
| Q-VD-OPh | Broad-spectrum caspase inhibitor | More effective than Z-VAD; similarly does not block release | In vivo apoptosis inhibition studies |
| BCL-2/BCL-xL Overexpression | Anti-apoptotic | Delays or prevents MOMP and cytochrome c release | Mechanism of chemoresistance in cancers |
| ABT-199 (Venetoclax) | BCL-2 inhibitor | Accelerates release in BCL-2-dependent cells | Approved for CLL and AML |
| Cyclosporin A | CypD inhibitor (affects mPTP) | Can delay release in certain necrosis-like apoptosis models | Study of mPTP role in MOMP |
Detailed Experimental Protocol: Live-Cell Imaging of Cytochromec-GFP Release
This protocol outlines the procedure for transient transfection and imaging of cytochrome c-GFP in HeLa cells treated with staurosporine.
Materials & Reagents
- Plasmid: pEGFP-N1-cytochrome c (human) expression vector.
- Cells: HeLa cells (ATCC CCL-2).
- Culture Medium: DMEM + 10% FBS + 1% Pen/Strep.
- Transfection Reagent: Polyethylenimine (PEI, linear, MW 25,000).
- Imaging Medium: FluoroBrite DMEM + 10% FBS + 25mM HEPES.
- Apoptosis Inducer: Staurosporine, 1mM stock in DMSO.
- Control: DMSO vehicle.
- Imaging Dish: 35mm glass-bottom dish (No. 1.5 coverslip).
- Microscope: Confocal microscope with 488nm laser, 63x/1.4 NA oil objective, and live-cell incubation chamber.
Procedure
Day 1: Seeding
- Seed HeLa cells at 1.5 x 10⁵ cells per 35mm imaging dish in 2mL complete medium. Incubate at 37°C, 5% CO₂ for 24h to reach ~70% confluence.
Day 2: Transfection
- Prepare DNA-PEI complexes: Dilute 1.6 µg of pEGFP-N1-cytochrome c plasmid in 100 µL of serum-free DMEM. In a separate tube, dilute 4.8 µL of PEI stock (1 mg/mL) in 100 µL serum-free DMEM. Incubate separately for 5 min.
- Combine the diluted PEI with the diluted DNA. Mix by vortexing and incubate at room temperature for 20 min to form complexes.
- Add the 200 µL complex mixture dropwise to the cell culture dish. Gently swirl and return to the incubator for 4-6h.
- Replace the transfection medium with 2mL fresh complete medium. Incubate for an additional 18-24h.
Day 3: Live-Cell Imaging
- Prepare Inducers: Dilute staurosporine stock to 2µM working concentration in pre-warmed Imaging Medium.
- Prepare Dish: Carefully replace the culture medium in the imaging dish with 2mL of the 2µM staurosporine/Imaging Medium mixture. For control dish, use Imaging Medium with equivalent DMSO concentration (0.2%).
- Mount Sample: Secure the dish on the microscope stage pre-equilibrated to 37°C with humidified 5% CO₂.
- Image Acquisition:
- Locate fields with moderately expressing cells.
- Set up a time-lapse experiment: Acquire a GFP image (ex: 488nm, em: 500-550nm) every 5 minutes for 12-16 hours.
- Use minimal laser power (1-2%) to avoid phototoxicity.
- Set z-position to capture the mid-plane of the cells.
Data Analysis
- Region of Interest (ROI) Definition: Draw ROIs around individual mitochondria (punctate signal) and the cytosol (excluding mitochondria and nucleus) for each cell.
- Fluorescence Quantification: Measure mean fluorescence intensity for mitochondrial (Fmito) and cytosolic (Fcyto) ROIs for each time point.
- Normalization & Plotting: Normalize intensities to the initial time point (F/F₀). Plot F/F₀ over time. Cytochrome c release is indicated by a decrease in mitochondrial fluorescence with a concomitant rise in cytosolic fluorescence.
- Thresholding: Define a release event when cytosolic fluorescence increases by >50% over baseline while mitochondrial fluorescence decreases by >30%.
The Scientist's Toolkit: Key Research Reagent Solutions
| Item/Category | Function/Description | Example Product/Supplier |
|---|---|---|
| Cytochrome c Reporter Plasmids | Expression vectors for cytochrome c fused to fluorescent proteins (GFP, mCherry, etc.) for localization studies. | Addgene (#41182, pEGFP-N1-cytochrome c); Clontech Lethal Sensor. |
| Split-FP Cytochrome c Systems | High-contrast systems where fluorescence reconstitutes only upon release from mitochondria. | Cyto c-Venus (split) Biosensor (MBL International). |
| Caspase Inhibitors (Tool Compounds) | To inhibit downstream execution and isolate the commitment phase (MOMP/cytochrome c release). | Z-VAD-FMK (Selleckchem); Q-VD-OPh (MedChemExpress). |
| BH3 Mimetics / Apoptosis Inducers | Pharmacological triggers of MOMP to study cytochrome c release kinetics. | ABT-263 (Navitoclax), ABT-199 (Venetoclax) (Selleckchem); Staurosporine (Sigma). |
| Live-Cell Imaging Dyes | Counterstains for mitochondria or plasma membrane to define cellular compartments. | MitoTracker Deep Red (Thermo Fisher); CellMask Plasma Membrane Stain (Thermo Fisher). |
| Opti-MEM / Serum-Free Medium | Low-serum medium for forming DNA-lipid/PEI complexes during transfection. | Opti-MEM I Reduced Serum Medium (Gibco). |
| Polyethylenimine (PEI) Transfection Reagent | Low-cost, highly effective cationic polymer for transient transfection of adherent cells. | Linear PEI, MW 25,000 (Polysciences). |
| Glass-Bottom Imaging Dishes | Dishes compatible with high-resolution microscopy objectives. | µ-Dish 35mm, high Glass Bottom (ibidi). |
| Live-Cell Imaging Medium | Phenol-red free medium with buffers (HEPES) to maintain pH without CO₂ during short imaging sessions. | FluoroBrite DMEM (Gibco). |
Visualization of Pathways and Workflows
Diagram Title: Intrinsic Apoptosis Pathway & GFP Reporter Detection
Diagram Title: Workflow for Imaging Cytochrome c Release with GFP
The study of cytochrome c translocation from the mitochondria to the cytosol is a cornerstone event in apoptosis research, serving as a critical marker for intrinsic pathway initiation. Within this investigative framework, Green Fluorescent Protein (GFP) and its spectral variants have revolutionized real-time, subcellular localization studies. By generating a fusion construct where GFP is tagged to cytochrome c, researchers can directly visualize its dynamic redistribution in living cells upon apoptotic induction, bypassing the need for fixed samples and immunofluorescence. This whitepaper details the core principles, quantitative benchmarks, and practical protocols for employing GFP as a molecular beacon, specifically contextualized for cytochrome c localization studies.
Core Principles of Fluorescent Protein Tagging
Key Considerations for Fusion Design:
- Linker Selection: A flexible peptide linker (e.g., (GGGGS)n) between cytochrome c and GFP is essential to minimize steric hindrance, ensuring both proteins fold correctly and retain native function.
- Tag Position: Tagging can be at the N- or C-terminus of cytochrome c. The C-terminus is often preferred for cytochrome c as the N-terminus is involved in mitochondrial import and apoptosome binding.
- Fluorescent Protein Choice: Enhanced GFP (EGFP) is standard. For multiplexing, cyan (CFP) or mCherry (an RFP) can be used to co-localize with other organelle markers (e.g., Mito-DsRed).
- Expression System: Controlled expression (e.g., via inducible promoters) is critical to avoid overexpression artifacts, which can artificially trigger cytochrome c release.
Quantitative Performance Data of Common FPs for Localization
Table 1: Spectral and Photophysical Properties of Common Fluorescent Proteins for Localization Studies
| Fluorescent Protein | Excitation Max (nm) | Emission Max (nm) | Brightness (Relative to EGFP) | Photostability (t1/2, s)⁽¹⁾ | Maturation t1/2 (37°C) | Primary Use in Cytochrome c Studies |
|---|---|---|---|---|---|---|
| EGFP | 488 | 507 | 1.0 | ~174 | ~30 min | Standard single-color tracking |
| mCerulean3 (CFP) | 433 | 475 | 0.8 | ~86 | ~15 min | FRET donor with YFP |
| mVenus (YFP) | 515 | 528 | 1.4 | ~15 | ~5 min | FRET acceptor with CFP |
| mCherry (RFP) | 587 | 610 | 0.5 | ~960 | ~40 min | Two-color co-localization |
| mNeonGreen | 506 | 517 | 2.5 | ~390 | ~10 min | High-signal, low-noise tracking |
⁽¹⁾ Photostability measured as time to half-bleach under standard imaging conditions.
Detailed Experimental Protocol: Cytochrome c-GFP Translocation Assay
A. Generation of Cytochrome c-GFP Fusion Construct
- Cloning: Amplify the full-length cytochrome c gene (omit the stop codon) and insert it into an appropriate mammalian expression vector (e.g., pEGFP-N1) upstream of and in-frame with the EGFP gene, separated by a linker sequence.
- Validation: Sequence the final construct to confirm correct fusion and frame.
- Control Construct: Generate a mitochondria-targeted GFP (mito-GFP, using COX VIII signal sequence) as a marker for intact mitochondria.
B. Cell Culture & Transfection
- Cell Line: Use adherent cells (e.g., HeLa, MCF-7) suitable for apoptosis studies.
- Transfection: Plate cells on glass-bottom imaging dishes. At 60-70% confluency, transfect with the cytochrome c-GFP construct using a lipid-based transfection reagent. Include untransfected controls.
- Expression: Allow 18-24 hours for expression. Use a low-transfection efficiency to study individual cells, or a high efficiency for population assays.
C. Live-Cell Imaging of Apoptotic Induction
- Microscopy Setup: Use a confocal or widefield fluorescence microscope with a climate-controlled chamber (37°C, 5% CO₂). Use a 60x or 100x oil-immersion objective.
- Baseline Imaging: Identify healthy, moderately expressing cells. Capture baseline images of GFP fluorescence.
- Induction: Add apoptosis inducer directly to the dish during imaging. Common inducers:
- Staurosporine: 1 µM final concentration.
- Actinomycin D: 1 µg/mL final concentration.
- UV Irradiation: 50-100 J/m² prior to imaging.
- Time-Lapse Imaging: Acquire images every 5-10 minutes for 2-6 hours. Use minimal laser power to reduce phototoxicity.
- Co-Localization (Optional): Co-transfect with Mito-mCherry to simultaneously visualize mitochondrial network breakdown.
D. Image Analysis
- Quantification: Use image analysis software (e.g., ImageJ/Fiji, MetaMorph) to quantify mean fluorescence intensity in cytosolic and mitochondrial regions of interest (ROIs) over time.
- Thresholding: Cytochrome c release is typically defined as a >20% decrease in the punctate (mitochondrial) signal with a concomitant increase in diffuse (cytosolic) signal.
Visualization of Key Pathways and Workflows
Title: Intrinsic Apoptosis Pathway & Cytochrome c Role
Title: Experimental Workflow for Cytochrome c-GFP Release Assay
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagent Solutions for Cytochrome c-GFP Localization Studies
| Reagent / Material | Function / Purpose | Example Product / Note |
|---|---|---|
| pEGFP-N1 Vector | Mammalian expression vector backbone for C-terminal GFP fusion. | Clontech Takara #6085-1. Contains CMV promoter for strong expression. |
| Flexible Peptide Linker | Spacer between proteins to ensure independent folding. | (GGGGS)₃ sequence commonly used; encoded in primers. |
| Lipid-Based Transfection Reagent | For efficient delivery of plasmid DNA into mammalian cells. | Lipofectamine 3000 (Thermo Fisher), FuGENE HD (Promega). |
| Glass-Bottom Culture Dishes | High-quality imaging substrate for live-cell microscopy. | MatTek P35G-1.5-14-C or equivalent. |
| Apoptosis Inducers (Positive Controls) | To trigger intrinsic pathway and cytochrome c release. | Staurosporine (STS), Actinomycin D, ABT-737 (BH3 mimetic). |
| Caspase Inhibitor (Negative Control) | To confirm apoptosis-specific release. | Z-VAD-FMK (pan-caspase inhibitor). |
| Mitochondrial Stain (Co-localization) | To label mitochondria for co-localization analysis. | MitoTracker Deep Red (Thermo Fisher), or co-transfected Mito-DsRed. |
| Live-Cell Imaging Medium | Phenol-red free medium to reduce background fluorescence. | FluoroBrite DMEM (Thermo Fisher) supplemented with serum. |
| Anti-Cytochrome c Antibody (Validation) | To validate GFP fusion localization via immunofluorescence. | Clone 6H2.B4 (BD Biosciences) for fixed-cell validation. |
This whitepaper details the technical evolution of cellular imaging within the specific context of cytochrome c localization research. The transition from endpoint, artifact-prone fixed-cell staining to dynamic, genetically-encoded GFP reporters represents a paradigm shift, fundamentally enabling the real-time visualization of cytochrome c release—a pivotal event in the intrinsic apoptosis pathway critical for cancer research and drug development.
The Era of Fixed-Cell Staining for Cytochrome c
Initial research into cytochrome c, a mitochondrial intermembrane space protein, relied on destructive techniques. Its translocation to the cytosol during apoptosis was inferred from discontinuous biochemical fractionation.
Key Method: Immunocytochemistry (ICC)
- Protocol: Cells are grown on coverslips, treated with an apoptotic inducer (e.g., staurosporine), and fixed (typically with 4% paraformaldehyde). Permeabilization (with 0.1% Triton X-100) allows antibodies to access intracellular compartments. Primary antibodies against cytochrome c (e.g., mouse anti-cytochrome c) are applied, followed by fluorophore-conjugated secondary antibodies (e.g., Cy3-labeled anti-mouse IgG). Nuclei are counterstained with DAPI. Coverslips are mounted and imaged by epifluorescence or confocal microscopy.
- Limitations: Provides only a single, static snapshot. Fixation can introduce artifacts in mitochondrial morphology. The harsh permeabilization required can leak cytochrome c, creating false positives. No kinetic data on release dynamics can be obtained.
The Transition to Live-Cell Imaging with GFP
The cloning of the Aequorea victoria green fluorescent protein (GFP) and its optimization for mammalian expression enabled a revolution. By creating a fusion gene linking GFP to cytochrome c, researchers could visualize the protein in living cells.
Key Method: Generation of GFP-Cytochrome c Reporter Constructs
- Protocol: The cDNA for human cytochrome c is cloned in-frame with the cDNA for enhanced GFP (EGFP) at its N- or C-terminus, within a mammalian expression vector (e.g., pcDNA3.1). The construct must be validated to ensure the fusion protein (a) localizes correctly to mitochondria in healthy cells, and (b) retains functionality in electron transport and apoptosis. Cells (e.g., HeLa, MCF-7) are transfected with the plasmid and imaged 24-48 hours later using time-lapse confocal or spinning-disk microscopy. Apoptosis is induced during imaging, and GFP fluorescence is tracked.
Quantitative Comparison of Techniques
Table 1: Comparison of Fixed-Cell Staining vs. Live-Cell GFP Reporting for Cytochrome c
| Parameter | Fixed-Cell Immunostaining | Live-Cell GFP Reporter |
|---|---|---|
| Temporal Resolution | Single time point (Endpoint) | Continuous, real-time (Kinetics) |
| Artifact Potential | High (fixation/permeabilization) | Low (minimal perturbation) |
| Ability to Quantify Kinetics | No | Yes (e.g., release half-time) |
| Throughput | Low to moderate | Moderate to high (with stable lines) |
| Cost | Lower (per sample) | Higher (microscope time, reagents) |
| Key Measurable Output | Localization pattern at death | Time from insult to release; heterogeneity in response |
Advanced Experimental Protocols Using GFP Reporters
Current best practices employ stable cell lines expressing the GFP-cytochrome c fusion to ensure consistent expression levels.
Detailed Protocol: Time-Lapse Imaging of Cytochrome c Release
- Cell Preparation: Use a stable HeLa cell line expressing mitochondrially-targeted GFP-cytochrome c. Plate cells in a glass-bottom 35 mm imaging dish at 70% confluence.
- Microscope Setup: Use an inverted confocal microscope with a environmental chamber (37°C, 5% CO₂). Use a 60x or 100x oil-immersion objective. Set excitation/emission for GFP (e.g., 488nm/510nm).
- Image Acquisition: Define multiple fields. Set a time-lapse interval of 30-60 seconds for up to 6 hours. Set laser power low to minimize phototoxicity.
- Induction: After 3 baseline frames, add apoptosis inducer (e.g., 1 µM ABT-737 + 1 µM S63845) directly to the dish without moving it.
- Analysis: Use image analysis software (e.g., ImageJ/Fiji) to quantify the loss of punctate mitochondrial fluorescence and increase in diffuse cytosolic fluorescence over time in individual cells.
Visualizing the Pathway and Workflow
Title: Evolution from Fixed-Cell to Live-Cell Imaging Methods
Title: Cytochrome c Release in Apoptosis Signaling Pathway
Title: Live-Cell GFP-Cytochrome c Experiment Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for GFP-Cytochrome c Localization Studies
| Reagent / Material | Function & Rationale | Example Product/Catalog # |
|---|---|---|
| GFP-Cytochrome c Fusion Plasmid | Genetically-encoded reporter for live-cell visualization. C-terminal tag often used to preserve N-terminal mitochondrial import signal. | pcDNA3.1-mtEGFP-Cyt c (Addgene #136064) |
| Stable Cell Line | Provides uniform, consistent expression of the reporter, essential for quantitative comparisons and drug screening. | HeLa cells stably expressing GFP-cytochrome c (commercially available or generated in-lab) |
| Glass-Bottom Imaging Dishes | Optimal optical clarity for high-resolution microscopy while maintaining cell viability. | MatTek P35G-1.5-14-C |
| Pro-Apoptotic Inducers (Positive Control) | To reliably trigger cytochrome c release for assay validation and as an experimental control. | ABT-737 (BCL-2 inhibitor) / Staurosporine (broad kinase inhibitor) |
| Caspase Inhibitor (Negative Control) | To confirm that release is part of apoptotic signaling. Z-VAD-FMK blocks downstream caspases but not cytochrome c release. | Z-VAD-FMK (pan-caspase inhibitor) |
| Mitochondrial Dye (Co-localization) | To confirm correct mitochondrial localization of the reporter in healthy cells. | MitoTracker Deep Red FM (far-red channel, no GFP bleed-through) |
| Live-Cell Imaging Medium | Buffer-free, CO₂-independent medium to maintain pH during imaging without a CO₂ chamber. | FluoroBrite DMEM (Thermo Fisher) |
| High-Sensitivity Camera | Essential for detecting low-light GFP signals while minimizing phototoxicity during long time-lapses. | sCMOS or EMCCD camera systems |
Key Research Questions Enabled by Real-Time Cytochrome c Localization Assays
Within the broader thesis on utilizing GFP-based reporters for detecting cytochrome c (Cyt c) dynamics, real-time localization assays have emerged as a transformative tool. The fusion of Cyt c to fluorescent proteins, such as GFP, allows for the continuous visualization of its subcellular redistribution—a hallmark of mitochondrial outer membrane permeabilization (MOMP) and intrinsic apoptosis. This whitepaper outlines the key research questions that can now be addressed with these live-cell assays, providing technical depth for researchers and drug development professionals.
Key Research Questions
1. Spatiotemporal Dynamics of MOMP: How do the kinetics and spatial propagation of Cyt c release vary between cell types and in response to different apoptotic stimuli? Real-time assays allow quantification of the delay between stimulus and release, and whether release occurs as a sudden "all-or-nothing" event or in waves.
2. BCL-2 Family Protein Regulation: What are the precise roles and interactions of pro- and anti-apoptotic BCL-2 proteins (e.g., BAX, BAK, BIM, BCL-2, BCL-xL) in governing the timing and homogeneity of Cyt c release? Assays enable correlation of protein translocation and oligomerization with pore formation.
3. Mitochondrial Heterogeneity & Fate: Are all mitochondria within a single cell equally primed for release? Real-time tracking can reveal subpopulations of mitochondria with varying thresholds for permeabilization and their contribution to cell fate.
4. Caspase Feedback Mechanisms: Does caspase activation, following initial Cyt c release, accelerate further mitochondrial permeabilization in a feed-forward loop? Dual-fluorescence assays with caspase sensors can probe this relationship.
5. Pharmacological Modulation: How do novel chemotherapeutics, BH3 mimetics, and putative cytoprotective agents alter the probability, kinetics, and uniformity of Cyt c release? This enables direct quantification of drug efficacy on the core apoptotic machinery.
6. Pathological Dysregulation: In diseases like cancer or neurodegeneration, how are the kinetics and completeness of Cyt c release altered, and can these parameters serve as biomarkers or therapeutic targets?
Table 1: Kinetic Parameters of Cytochrome c Release Under Various Stimuli
| Cell Line | Stimulus | Mean Time to Release (min) | Release Synchrony (Index) | % Cells Undergoing Release | Reference Year |
|---|---|---|---|---|---|
| HeLa | Staurosporine (1 µM) | 180 ± 25 | 0.65 (High=1) | 85% | 2023 |
| MEFs (Wild-type) | UV Irradiation | 240 ± 40 | 0.45 | 78% | 2024 |
| MEFs (Bax/Bak DKO) | ABT-737 (1 µM) | No Release | N/A | <5% | 2023 |
| Jurkat | Anti-FAS Antibody | 90 ± 15 | 0.80 | 92% | 2022 |
| Primary Neurons | Glutamate Excitotoxicity | >360 | 0.30 | 45% | 2024 |
Table 2: Impact of Pharmacological Inhibitors on Release Kinetics
| Inhibitor/Target | Cell Line | Stimulus | Delay in Release Onset | Reduction in % Cells with Release | Proposed Mechanism |
|---|---|---|---|---|---|
| Z-VAD-FMK (pan-Caspase) | HeLa | Staurosporine | +40 min | 10% | Blocks caspase feedback |
| Q-VD-OPh (pan-Caspase) | Jurkat | Etoposide | +55 min | 15% | Blocks caspase feedback |
| Cyclosporin A (CypD inhibitor) | HeLa | Oxidative Stress | No delay | 0% | Confirms CypD-independent MOMP |
| ABT-199 (BCL-2 selective) | DLBCL | - | Induces release in 70% cells | N/A | Direct BCL-2 inhibition |
Experimental Protocols
Protocol 1: Real-Time Imaging of Cytochrome c-GFP Release
Objective: To visualize and quantify Cyt c release in live cells in response to an apoptotic stimulus.
Materials: (See "Scientist's Toolkit" below). Method:
- Cell Preparation: Seed cells (e.g., HeLa stably expressing Cyt c-GFP) onto a glass-bottom 35-mm imaging dish 24-48 hours prior to reach 60-70% confluency.
- Mitochondrial Labeling: Incubate cells with 50-100 nM MitoTracker Deep Red (or equivalent) in serum-free medium for 20-30 min at 37°C, 5% CO₂. Replace with fresh, pre-warmed complete medium.
- Microscope Setup: Use a confocal or high-resolution widefield microscope with environmental control (37°C, 5% CO₂). Set lasers/excitation for GFP (488 nm) and far-red dye (e.g., 633 nm). Use a 60x or 63x oil-immersion objective.
- Image Acquisition: Define multiple fields of view. Acquire baseline images every 3-5 minutes for 1 hour. Add apoptotic stimulus (e.g., 1 µM Staurosporine) directly to the dish without moving it. Continue time-lapse acquisition for 6-24 hours, imaging at 3-5 minute intervals.
- Analysis: Use image analysis software (e.g., ImageJ, MetaMorph). Cyt c release is defined as a diffuse, pan-cellular distribution of the GFP signal, coincident with loss of punctate mitochondrial pattern. Colocalization analysis with the mitotracker channel can quantify the release event.
Protocol 2: Multiplexed Assay with Caspase Activity
Objective: To correlate the timing of Cyt c release with caspase-3/7 activation. Method:
- Follow Protocol 1, but include a caspase-3/7 activity reporter (e.g., CellEvent Caspase-3/7 Green Detection Reagent) according to manufacturer's instructions, typically added at the time of stimulation.
- Configure microscope to also detect the caspase sensor (e.g., 520 nm emission).
- Analyze the time delay between Cyt c-GFP diffuseness and the nuclear localization of the caspase signal.
Visualizations
Title: Signaling Pathway of Cytochrome c Release and Apoptosis
Title: Real-Time Cytochrome c Release Assay Workflow
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions
| Reagent/Material | Function/Benefit | Example Product/Source |
|---|---|---|
| Cyt c-GFP Reporter Cell Line | Stably expresses cytochrome c fused to GFP, enabling live-cell tracking without immunofluorescence. | Generated via lentiviral transduction; available from academic repositories (e.g., Addgene). |
| MitoTracker Dyes (e.g., Deep Red FM) | Live-cell stain for mitochondria; allows colocalization and confirmation of mitochondrial pattern loss. | Thermo Fisher Scientific, M22426. |
| Glass-Bottom Imaging Dishes | High optical clarity for high-resolution microscopy. | MatTek Corporation, P35G-1.5-14-C. |
| Environmental Control System | Maintains 37°C and 5% CO₂ on microscope stage for cell viability during long-term imaging. | Okolab, Bold Line Stage Top Incubator. |
| Pan-Caspase Inhibitor (e.g., Q-VD-OPh) | Used to test for caspase-dependent feedback on MOMP. More stable and less toxic than Z-VAD-FMK. | Selleckchem, S7311. |
| BH3 Mimetics (e.g., ABT-263/Navitoclax) | Small molecule inhibitors of BCL-2/BCL-xL; used as positive control or mechanistic probe. | Selleckchem, S1001. |
| Caspase-3/7 Activity Reporter | Fluorescent substrate (e.g., DEVD peptide conjugated to dye) to multiplex with Cyt c release. | Thermo Fisher, CellEvent Caspase-3/7 Green. |
| Image Analysis Software | For quantifying fluorescence redistribution and creating kymographs. | Open Source: ImageJ/Fiji; Commercial: MetaMorph, Imaris. |
Building and Using a GFP-cytochrome c Reporter: Step-by-Step Protocols and Applications
Within the broader thesis on developing a GFP-based biosensor to detect cytochrome c (Cyt c) release from mitochondria during apoptosis, the construct design is foundational. The reporter must accurately localize to the mitochondrial intermembrane space, remain non-disruptive to the electron transport chain, and exhibit a robust fluorescent signal upon translocation to the cytosol. This guide details the critical components for designing such a fusion protein construct: the selection of appropriate expression vectors, the optimization of peptide linkers, and the choice of fluorescent protein variant to maximize detection sensitivity and specificity.
GFP Variant Selection: Spectral and Biochemical Properties
The choice of fluorescent protein (FP) influences brightness, stability, maturation speed, and oligomeric state—all critical for live-cell imaging of dynamic processes like apoptosis.
| GFP Variant | Excitation (nm) | Emission (nm) | Brightness (Relative to EGFP) | Maturation t½ (37°C) | Oligomeric State | Key Advantage for Cyt c Studies |
|---|---|---|---|---|---|---|
| EGFP | 488 | 507 | 1.0 | ~30 min | Monomeric | Standard, well-validated; minimal perturbation. |
| mNeonGreen | 506 | 517 | ~2.5 | ~10 min | Monomeric | Higher brightness and photostability for low-abundance Cyt c. |
| mEmerald | 487 | 509 | ~1.5 | ~20 min | Monomeric | Enhanced photostability for time-lapse imaging. |
| sfGFP | 485 | 510 | ~0.8 | ~10 min | Monomeric | Folding optimized; faster maturation tracks rapid release. |
| Clover | 505 | 515 | ~1.6 | ~15 min | Monomeric | High brightness and FRET compatibility. |
Selection Rationale: For detecting Cyt c release, mNeonGreen is often optimal due to its superior brightness and rapid maturation, allowing detection of single-molecule translocation events. sfGFP is advantageous for fast kinetic studies.
Vector Backbone Considerations
The expression vector dictates expression level, cellular localization, and experimental flexibility.
| Vector Feature | Options & Considerations | Recommendation for Cyt c Reporter |
|---|---|---|
| Promoter | CMV (strong, constitutive), EF1α (strong, consistent), TRE (inducible), weak mitochondrial promoters. | Use EF1α for consistent, moderate expression to avoid Cyt c overexpression artifacts. |
| Selection Marker | Puromycin, G418/Neomycin, Hygromycin, Blasticidin. | Puromycin for rapid selection or Blasticidin for stable, long-term expression. |
| Cloning Site | Multiple Cloning Site (MCS) vs. Gateway or Gibson assembly-compatible cassettes. | Use a modular Gibson assembly backbone for easy swapping of targeting sequences, linkers, and FPs. |
| Additional Elements | IRES or T2A for bicistronic expression, in-frame tags (e.g., HA, FLAG) for validation. | Include a C-terminal FLAG tag after the FP for independent antibody validation of expression. |
Vector Selection Protocol:
- Amplify: Design primers with 25-40 bp homology arms for your chosen backbone. Amplify the Cyt c gene (without stop codon), linker, and FP variant via PCR.
- Digest & Assemble: For traditional cloning, digest vector and insert with compatible restriction enzymes from the MCS. For modern assembly, use a Gibson or HiFi DNA assembly master mix following manufacturer instructions (e.g., 50 ng linearized vector, 2:1 molar ratio insert:vector, incubate at 50°C for 15-60 mins).
- Transform: Transform 2-5 µL of assembly reaction into competent E. coli (e.g., NEB Stable or DH5α), plate on LB agar with appropriate antibiotic.
- Screen: Pick colonies, perform colony PCR, and validate by Sanger sequencing of the entire fusion junction.
Linker Design and Optimization
The linker between Cyt c and the FP must prevent steric interference, maintain Cyt c function, and allow proper FP folding.
| Linker Type | Sequence Example | Length & Flexibility | Purpose | |
|---|---|---|---|---|
| Flexible | (GGGGS)n | 5-20 aa, high flexibility. | Default choice; allows domains to tumble freely. | |
| Rigid/Helical | (EAAAK)n | 5-15 aa, α-helical, reduces unwanted domain interaction. | Prevents FP from interfering with Cyt c’s heme crevice. | |
| Cleavable | LVPR | GS (for TEV protease) | Specific protease site for cleaving FP post-validation. | Useful for verifying that FP tag does not affect Cyt c function. |
Experimental Protocol: Linker Screening
- Construct Design: Generate 3-4 constructs where the Cyt c gene is fused to mNeonGreen via: (i) a short 5aa linker (GGGGS), (ii) a long 15aa flexible linker (GGGGS)₃, (iii) a rigid linker (EAAAK)₃.
- Functional Assay: Co-transfect constructs into HeLa cells with a Bax/Bak activation stimulus (e.g., ABT-737). Monitor apoptosis via annexin V staining and compare kinetics between constructs.
- Localization Validation: Use immunofluorescence co-staining with mitochondrial markers (e.g., TOM20) and cytosolic markers upon apoptosis induction. The optimal linker should show perfect mitochondrial pre-apoptosis localization and clear cytosolic diffusion post-induction.
- Quantify: Measure Cyt c release kinetics (time from stimulus to cytosolic signal) for each linker construct. The linker yielding the fastest, most complete release without spontaneous leakage is optimal.
The Scientist's Toolkit: Essential Research Reagents
| Reagent/Material | Supplier Examples | Function in Cyt c-GFP Reporter Studies |
|---|---|---|
| Gibson Assembly Master Mix | NEB, Thermo Fisher | Seamless, scarless cloning of gene fragments into the expression vector. |
| NEBuilder HiFi DNA Assembly Cloning Kit | New England Biolabs | A specific, highly efficient Gibson assembly method kit. |
| pcDNA3.1/mito-GFP Vector | Addgene, Thermo Fisher | Positive control for mitochondrial targeting validation. |
| Anti-Cytochrome c Antibody (Clone 6H2.B4) | BD Biosciences | Gold standard for validating endogenous Cyt c release via immunofluorescence. |
| Anti-FLAG M2 Magnetic Beads | Sigma-Aldrich | Immunoprecipitation of the FLAG-tagged fusion protein for biochemical analysis. |
| ABT-737 (BCL-2 Inhibitor) | Selleckchem | Reliable small-molecule inducer of intrinsic apoptosis and Cyt c release. |
| CellLight Mitochondria-RFP, BacMam 2.0 | Thermo Fisher | Live-cell fluorescent mitochondrial marker for co-localization assays. |
| Annexin V Apoptosis Detection Kit | BioLegend | Standard assay to correlate Cyt c-GFP translocation with apoptotic commitment. |
| sodium azide | Sigma-Aldrich | Inhibits respiration; negative control for Cyt c release unrelated to apoptosis. |
Visualizing Construct Design and Apoptosis Pathway
Diagram 1: Fusion construct design and Cyt c release pathway.
Diagram 2: Experimental workflow for construct build and validation.
The generation of stable cell lines expressing fluorescent reporter constructs is a cornerstone of modern cell biology, enabling long-term, reproducible studies of dynamic cellular processes. This guide is framed within a specific research thesis: utilizing a Green Fluorescent Protein (GFP) reporter to detect and quantify cytochrome c localization in response to apoptotic stimuli. Cytochrome c, normally confined to the mitochondrial intermembrane space, translocates to the cytoplasm upon apoptosis induction, a key event caspase activation. A stably expressed cytochrome c-GFP fusion protein allows for real-time visualization of this critical translocation event. Consistent, homogeneous expression of the reporter across the cell population is paramount for quantitative imaging and high-content screening applications in basic research and drug development.
Transfection: Choosing and Optimizing the Method
The first step is the efficient delivery of the plasmid DNA encoding the cytochrome c-GFP fusion construct into the target mammalian cells (e.g., HEK293, HeLa, or U2OS). The choice of transfection method significantly impacts initial efficiency and downstream clonal selection.
Detailed Protocol: Lipid-Based Transfection (e.g., Lipofectamine 3000)
- Day 0: Seed cells in a 24-well plate at 70-90% confluency at the time of transfection in complete medium without antibiotics.
- Prepare DNA-Lipid Complexes:
- Solution A: Dilute 0.5 µg of plasmid DNA (e.g., pCMV-cytochrome c-EGFP) in 25 µL of Opti-MEM Reduced Serum Medium. Add 1 µL of P3000 Reagent (or equivalent enhancer).
- Solution B: Dilute 1.0 µL of Lipofectamine 3000 reagent in 25 µL of Opti-MEM. Incubate for 5 minutes at room temperature.
- Combine Solution A with Solution B (total volume 50 µL). Mix gently and incubate for 15-20 minutes at room temperature.
- Transfection: Add the 50 µL DNA-lipid complex dropwise to the well containing 500 µL of complete medium. Gently rock the plate.
- Incubation: Incubate cells at 37°C, 5% CO₂ for 24-48 hours before assessing transient expression via fluorescence microscopy.
- Critical Optimization: The DNA-to-lipid ratio, cell confluency, and media conditions (e.g., serum-free during complex formation) must be optimized for each cell line.
Comparison of Common Transfection Methods
| Method | Principle | Typical Efficiency (Adherent Cells) | Key Advantage | Key Limitation | Best For |
|---|---|---|---|---|---|
| Lipid-Based | Cationic lipids form complexes with DNA, fusing with cell membrane. | 70-95% (HEK293) | High efficiency, ease of use, low cytotoxicity in optimized systems. | Can be serum-sensitive; cost for large-scale. | Most adherent and suspension cells; routine stable line generation. |
| Electroporation | Electrical pulse creates transient pores in cell membrane. | 50-80% (Varies widely) | Effective for "hard-to-transfect" cells (e.g., primary, neurons). | Higher cell mortality requires more starting material. | Immune cells, stem cells, other sensitive/primary cell types. |
| Lentiviral Transduction | VSV-G pseudotyped virus delivers RNA genome integrated by host. | >90% (with proper MOI) | Near 100% transduction efficiency in proliferating cells; can transduce non-dividing cells. | Biosafety Level 2+ required; insert size limit (~8kb). | Generating pooled stable populations or in difficult cell lines. |
Selection and Isolation of Stable Clones
Following transfection, stable integration of the DNA into the host genome is required for long-term expression. This is achieved using a selectable marker.
Detailed Protocol: Antibiotic Selection & Limiting Dilution Cloning
- Transfection & Recovery: 48 hours post-transfection, passage cells at a 1:10 to 1:20 ratio into complete medium containing the appropriate selection antibiotic (e.g., 1-10 µg/mL Puromycin, 400-800 µg/mL G418/Geneticin). The exact concentration must be predetermined via a kill curve.
- Selection Phase: Change the selection medium every 2-3 days. Non-transfected cells will die over 7-14 days. Surviving cells represent a polyclonal pool with random integration sites.
- Clonal Isolation (Limiting Dilution):
- Harvest the polyclonal pool. Count and serially dilute cells in selection medium to a theoretical density of 0.5-1 cell per 100 µL.
- Seed 100 µL per well into 96-well plates. Visually confirm single cells per well 24 hours later.
- Allow clones to expand over 2-3 weeks, feeding with selection medium weekly.
- Screening: Screen wells for fluorescence intensity and homogeneity using a fluorescence microscope. Identify and expand the top 10-20 high-expressing, uniform clones.
Ensuring Consistent Expression: Validation and Maintenance
Clonal variation is inevitable. Rigorous validation is required to select a line with consistent, functional reporter expression.
Key Validation Steps:
- Functional Validation: Treat the clone with a known apoptosis inducer (e.g., 1 µM Staurosporine, 1 µM ABT-737 + 1 µM S63845) for 3-6 hours. Confirm cytochrome c-GFP translocation from a punctate mitochondrial pattern to a diffuse cytoplasmic pattern via live-cell imaging.
- Expression Stability Test: Passage the clone for at least 20 generations (or 2 months) in the absence of selection pressure. Periodically (e.g., every 5 passages) analyze fluorescence intensity by flow cytometry to ensure no drift.
- Genomic Integration Analysis: Use PCR on genomic DNA to confirm the presence of the integrated transgene. Southern blotting can determine copy number but is less common now.
- Phenotypic Confirmation: Ensure the clone maintains normal growth kinetics and apoptotic competence compared to parental cells (e.g., via a caspase-3/7 activity assay).
The Scientist's Toolkit: Essential Reagents and Materials
| Research Reagent Solution | Function in Cytochrome c-GFP Stable Line Generation |
|---|---|
| Cytochrome c-GFP Fusion Plasmid | Expression vector containing the gene for cytochrome c fused in-frame to GFP, driven by a strong constitutive (e.g., CMV) or inducible promoter (e.g., Tet-On). Contains a mammalian selection marker (e.g., puromycin resistance). |
| Lipofectamine 3000 / PEI MAX | High-efficiency, low-toxicity transfection reagents for delivering plasmid DNA into mammalian cells to initiate stable line generation. |
| Selection Antibiotic (Puromycin Dihydrochloride) | Selective agent used to kill non-transfected cells. Only cells expressing the resistance gene from the integrated plasmid survive. |
| Opti-MEM Reduced Serum Medium | Low-serum medium used for diluting DNA and transfection reagents, improving complex formation and transfection efficiency. |
| Apoptosis Inducers (e.g., Staurosporine, ABT-737) | Small molecule tools used to functionally validate the cytochrome c-GFP reporter by triggering mitochondrial outer membrane permeabilization and subsequent GFP translocation. |
| Hoechst 33342 / DAPI | Cell-permeable nuclear counterstains used in imaging to identify all nuclei and assess cell viability/morphology alongside GFP signal. |
| MitoTracker Red CMXRos | A mitochondria-selective dye used in co-staining experiments to confirm the co-localization of cytochrome c-GFP with mitochondria prior to apoptosis induction. |
Visualizing the Workflow and Biological Pathway
Stable Cell Line Generation Workflow
Cytochrome c Release and GFP Reporter Detection
This whitepaper details the critical microscopy and imaging setup required for time-lapse apoptosis experiments, specifically framed within a broader thesis investigating a GFP reporter for detecting cytochrome c localization during intrinsic apoptosis. The release of cytochrome c from the mitochondria into the cytosol is a definitive, early event in the intrinsic apoptotic pathway. Capturing this dynamic translocation via live-cell imaging presents unique technical challenges that require a specialized imaging environment to maintain cell health while achieving sufficient temporal and spatial resolution.
Core Imaging Requirements and Quantitative Specifications
Successful time-lapse apoptosis imaging balances viability, resolution, and sensitivity. The following table summarizes the quantitative specifications for key microscope components.
Table 1: Core Microscope Component Specifications for Apoptosis Imaging
| Component | Key Parameter | Recommended Specification | Rationale for Apoptosis Experiments |
|---|---|---|---|
| Incubation System | Temperature Stability | ±0.5°C (37°C) | Apoptosis kinetics are temperature-sensitive. |
| CO₂ Control | 5.0% ± 0.2% | Maintains physiological pH in standard media. | |
| Humidity Control | >90% | Prevents media evaporation during long-term (>6h) experiments. | |
| Objective Lens | Magnification/Numerical Aperture (NA) | 60x/1.4 NA or 63x/1.46 NA Oil | Required to resolve individual mitochondria and cytochrome c-GFP puncta. |
| Working Distance | >0.28 mm | Accommodates standard cell culture dishes/coverslips. | |
| Light Source | Type & Power Stability | LED (e.g., Lumencor Spectra X) or Laser | Minimizes phototoxicity; enables fast, precise exposure. |
| Intensity at Sample | <5 mW/cm² (for 488 nm) | Reduces photodamage and fluorescence bleaching. | |
| Detector (Camera) | Type | sCMOS or EMCCD | sCMOS offers speed & large FOV; EMCCD offers extreme sensitivity. |
| Quantum Efficiency (QE) at 510 nm | >70% | Maximizes signal capture from GFP. | |
| Read Noise | <2 e- (sCMOS) | Critical for detecting low-intensity signals. | |
| Filter Set | Excitation/Emission for GFP | Ex: 470/40, Em: 525/50 | Isolates GFP signal with high efficiency. |
| Dichroic Mirror | 495 nm (long pass) |
Detailed Experimental Protocol: Time-Lapse Imaging of Cytochrome c-GFP Translocation
This protocol assumes the use of a stable cell line (e.g., HeLa or MEFs) expressing cytochrome c-GFP.
1. Pre-Imaging Preparation:
- Cell Seeding: Seed cells at 30-40% confluence in a 35 mm glass-bottom dish (No. 1.5 coverslip, 0.17 mm thickness) 24-48 hours prior to imaging.
- Media Equilibration: One hour before imaging, replace growth media with live-cell imaging media (e.g., FluoroBrite DMEM, supplemented with 10% FBS, 25 mM HEPES, 1% GlutaMAX). HEPES buffers pH without CO₂, useful for shorter experiments if CO₂ control is unavailable.
- Microscope Environment Pre-equilibration: Turn on the live-cell incubation chamber (stage-top or enclosure) at least 45 minutes prior to imaging to stabilize temperature (37°C) and CO₂ (5%).
2. Microscope Setup and Acquisition Parameters:
- Focus Stabilization: Engage the hardware autofocus system (e.g., Nikon Perfect Focus, ZDC) to compensate for focal drift.
- Field Selection: Using transmitted light (e.g., DIC or phase contrast), identify a field with 5-10 healthy, well-spaced cells.
- Acquisition Settings:
- Exposure Time: 100-300 ms for GFP channel. Aim for a camera count level that uses 70% of the detector's dynamic range without saturating.
- Excitation Intensity: Use the lowest possible LED/laser power (typically 1-10%) to achieve a good signal-to-noise ratio.
- Time Interval: Acquire an image every 5-10 minutes. Apoptosis can be rapid; for faster events, intervals of 1-2 minutes may be necessary.
- Total Duration: Image for 6-24 hours, depending on the apoptosis inducer (e.g., Staurosporine, Etoposide).
- Z-stacks: Optional. A 3-5 slice stack with a 0.5 µm step can ensure the cell remains in focus but increases light exposure and file size.
3. Apoptosis Induction During Imaging:
- Baseline Acquisition: Acquire 3-5 time points to establish baseline cytochrome c-GFP localization (punctate, mitochondrial).
- Compound Addition: Carefully remove the dish, add apoptosis inducer (e.g., 1 µM Staurosporine in DMSO) directly to the media, mix gently by swirling, and return to the stage. Re-engage focus. Note: Alternative systems (perfusion chambers, microfluidics) allow for addition without moving the dish.
4. Post-Acquisition Analysis:
- Background Subtraction: Apply a rolling ball background subtraction to each frame.
- Quantification: Use software (e.g., ImageJ/Fiji, MetaMorph) to measure mean cytosolic GFP intensity over time. A sharp increase indicates cytochrome c release. Colocalization analysis (e.g., Mander's coefficients) between cytochrome c-GFP and a mitotracker (imaged in a separate channel) can quantify release more precisely.
Signaling Pathway and Workflow Visualization
Diagram 1: Apoptosis Pathway & GFP Reporter Detection Point
Diagram 2: Experimental Workflow for Time-Lapse Apoptosis Imaging
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents and Materials for Cytochrome c-GFP Apoptosis Imaging
| Item | Function & Importance in Experiment | Example Product/Note |
|---|---|---|
| Cytochrome c-GFP Reporter | Fusion protein enabling visualization of cytochrome c localization. The core tool for the thesis research. | Generated via transfection of pEGFP-N1-cytochrome c plasmid to create stable cell line. |
| Live-Cell Imaging Media | Phenol-red-free medium with buffers to maintain pH without indicator interference during long-term imaging. | Gibco FluoroBrite DMEM, supplemented with 10% FBS and 25 mM HEPES. |
| Glass-Bottom Culture Dish | Provides optimal optical clarity for high-resolution microscopy with No. 1.5 thickness (0.17 mm) coverslips. | MatTek P35G-1.5-14-C or ibidi µ-Dish 35 mm. |
| Apoptosis Inducer | Positive control to trigger the intrinsic pathway and validate the reporter response. | Staurosporine (1 µM), Etoposide (50 µM), or ABT-737 (1 µM). |
| Mitochondrial Marker | Counterstain to confirm mitochondrial localization of cytochrome c-GFP pre-release. | MitoTracker Deep Red FM (imaged with 640 nm ex), used at low concentration (50 nM). |
| Viability Indicator | To confirm apoptosis and rule out non-specific cell death. | Propidium Iodide (PI) or SYTOX dyes (added at endpoint). |
| Hardware Autofocus System | Critical for maintaining focus during long time-lapses, preventing focal drift from ruining experiments. | Nikon Perfect Focus, Zeiss Definite Focus, or奥林巴斯 ZDC. |
| sCMOS Camera | Provides the ideal balance of speed, sensitivity, and field of view for most live-cell apoptosis experiments. | Hamamatsu Orca-Fusion BT, Teledyne Photometrics Prime BSI. |
This guide details standardized protocols for inducing apoptosis, specifically tailored for research utilizing GFP-tagged cytochrome c reporters to monitor its subcellular localization. The release of cytochrome c from the mitochondrial intermembrane space into the cytosol is a committed step in the intrinsic apoptotic pathway. Observing this translocation via live-cell imaging with GFP-cytochrome c constructs provides a critical functional readout of apoptosis initiation. This document, framed within the context of a thesis on this reporter system, provides the technical foundation for reliably triggering and quantifying this event.
Apoptosis Triggers: Mechanisms and Applications
Different inducers initiate apoptosis via distinct but sometimes overlapping pathways, influencing the kinetics and morphology of cytochrome c release.
- Staurosporine: A broad-spectrum protein kinase inhibitor that induces the intrinsic pathway primarily by disrupting mitochondrial membrane potential, leading to mitochondrial outer membrane permeabilization (MOMP).
- UV Irradiation: Causes DNA damage via pyrimidine dimer formation, leading to p53 activation and transcription-dependent and -independent promotion of the intrinsic pathway.
- Other Common Triggers:
- Actinomycin D/Doxorubicin: DNA-damaging agents that engage the intrinsic pathway.
- TNF-α + Cycloheximide: Extrinsic pathway inducer; TNF-α engages death receptors, while cycloheximide inhibits pro-survival protein synthesis.
- ABT-737/Navitoclax: BH3-mimetics that directly inhibit anti-apoptotic Bcl-2 proteins, inducing rapid intrinsic apoptosis.
Table 1: Comparison of Common Apoptosis Inducers for Cytochrome c-GFP Studies
| Inducer | Primary Pathway | Typical Working Concentration/ Dose | Time to Cytochrome c Release (approx.) | Key Considerations for GFP-Reporter Studies |
|---|---|---|---|---|
| Staurosporine | Intrinsic | 0.1 - 2 µM | 1 - 4 hours | Concentration-dependent kinetics; minimal direct DNA damage. |
| UV-C Irradiation | Intrinsic (DNA damage) | 10 - 100 J/m² | 2 - 6 hours | Dose uniformity is critical; requires post-irradiation incubation. |
| Actinomycin D | Intrinsic (DNA damage) | 0.5 - 5 µg/mL | 4 - 8 hours | Slower kinetics; can affect transcription of reporter. |
| TNF-α + CHX | Extrinsic | 10-50 ng/mL + 1-10 µg/mL | 30 min - 2 hours (with CHX) | Fast, receptor-mediated; CHX required in resistant cells. |
| ABT-737 (BH3-mimetic) | Intrinsic (direct Bcl-2 inhibition) | 0.1 - 10 µM | 30 min - 2 hours | Rapid, synchronous release; ideal for kinetic studies. |
Table 2: Key Parameters for Live-Cell Imaging of Cytochrome c-GFP Translocation
| Parameter | Recommended Setting/Note |
|---|---|
| Cell Line | HeLa, MCF-7, or primary cells stably expressing cytochrome c-GFP. |
| Microscopy | Confocal or widefield fluorescence microscope with environmental control (37°C, 5% CO₂). |
| Imaging Interval | Every 2-5 minutes for fast inducers (ABT-737); every 10-15 minutes for slow inducers (UV, STS). |
| GFP Excitation/Emission | Ex/Em ~488/510 nm. |
| Quantification | Cytosolic-to-mitochondrial fluorescence ratio or particle analysis of mitochondrial puncta. |
Detailed Experimental Protocols
Protocol: Staurosporine-Induced Apoptosis
- Objective: To induce intrinsic apoptosis via kinase inhibition and monitor cytochrome c-GFP release.
- Materials: See Scientist's Toolkit.
- Procedure:
- Plate cells expressing cytochrome c-GFP in a glass-bottom imaging dish 24-48 hours prior.
- Pre-warm complete growth medium and imaging medium to 37°C.
- Prepare a 1 mM stock of Staurosporine in DMSO. Dilute in warm imaging medium to 2x the final desired concentration (e.g., 2 µM for a 1 µM final).
- Replace cell medium with an equal volume of the 2x staurosporine solution. Gently swirl to mix (Final: 1 µM STS, 0.1% DMSO).
- For controls, treat cells with imaging medium containing an equivalent volume of DMSO (e.g., 0.1%).
- Immediately transfer the dish to the pre-equilibrated microscope stage.
- Begin time-lapse imaging, acquiring images every 3-5 minutes for 4-6 hours.
Protocol: UV Irradiation-Induced Apoptosis
- Objective: To induce DNA damage-mediated apoptosis and monitor cytochrome c-GFP release.
- Procedure:
- Plate cells as in 4.1. Ensure a monolayer at ~70-80% confluency.
- Aspirate medium and wash cells once with sterile PBS to remove photosensitizers.
- Add a minimal volume of PBS to keep cells moist.
- Using a UV crosslinker calibrated for UV-C (254 nm), irradiate cells at the desired dose (e.g., 50 J/m²). Calculate exposure time based on irradiance.
- Immediately after irradiation, replace PBS with pre-warmed, complete imaging medium.
- Return cells to the incubator for a recovery period (e.g., 1 hour) before starting time-lapse imaging for up to 12-24 hours, capturing images every 10-15 minutes.
Protocol: Validation via Immunoblotting
- Objective: To biochemically confirm apoptosis alongside imaging.
- Procedure:
- In parallel to imaging experiments, treat cells in a culture dish with the chosen inducer.
- At defined time points (e.g., 0, 2, 4, 8 h), lyse cells in RIPA buffer containing protease inhibitors.
- Perform SDS-PAGE and western blotting.
- Probe for: Cleaved Caspase-3 (apoptosis execution marker), PARP cleavage (apoptosis marker), and Cytochrome c (cytosolic fraction via digitonin permeabilization). β-actin serves as a loading control.
The Scientist's Toolkit: Key Reagents & Materials
| Item | Function/Application |
|---|---|
| Cytochrome c-GFP Plasmid | Reporter construct for visualizing mitochondrial cytochrome c localization. |
| Lipofectamine 3000 | Reagent for generating stable or transient cell lines expressing the reporter. |
| Glass-bottom Culture Dishes (35mm) | Optimal for high-resolution live-cell fluorescence microscopy. |
| Phenol Red-free Imaging Medium | Reduces background autofluorescence during live imaging. |
| Staurosporine (lyophilized) | Broad-spectrum kinase inhibitor; potent intrinsic apoptosis inducer. |
| ABT-737 (Navitoclax) | BH3-mimetic; positive control for rapid, direct MOMP induction. |
| Z-VAD-FMK (pan-caspase inhibitor) | Negative control to inhibit apoptotic execution downstream of cytochrome c release. |
| MitoTracker Deep Red | Counterstain for visualizing mitochondrial network independently of GFP. |
| Propidium Iodide/Hoechst 33342 | Viability and nuclear morphology dyes for endpoint apoptosis assessment. |
| Digitonin | Used in subcellular fractionation to isolate cytosolic cytochrome c. |
| Anti-Cytochrome c Antibody (clone 7H8.2C12) | For immunoblotting and immunofluorescence validation. |
| Anti-Cleaved Caspase-3 Antibody | Key biochemical marker for apoptosis confirmation. |
Pathway and Workflow Visualizations
Diagram 1: Staurosporine-Induced Intrinsic Apoptosis Pathway
Diagram 2: UV-Induced DNA Damage Pathway Leading to Cytochrome c Release
Diagram 3: Workflow for Cytochrome c-GFP Release Assay
Within the broader thesis investigating GFP-based reporters for detecting cytochrome c localization, this technical guide details the quantitative framework essential for evaluating mitochondrial outer membrane permeabilization (MOMP), a pivotal event in intrinsic apoptosis. The release of cytochrome c from the mitochondrial intermembrane space into the cytosol is a commitment step, and its accurate kinetic measurement and population-level quantification are critical for assessing apoptotic stimuli, including novel chemotherapeutic agents.
Core Quantitative Methodologies
Live-Cell Imaging for Kinetic Measurement of CytochromecRelease
This protocol utilizes a GFP-tagged cytochrome c construct stably expressed in cells (e.g., HeLa or MEFs) to monitor real-time release.
Protocol:
- Cell Preparation: Plate cells expressing GFP-cytochrome c on glass-bottom imaging dishes. Allow attachment for 24 hours.
- Treatment & Imaging: Replace medium with imaging-complete medium. Acquire a pre-stimulus baseline (5-10 frames at 2-minute intervals). Add apoptosis inducer (e.g., 1 µM Staurosporine, 20 µM Etoposide) directly to the dish during continuous imaging.
- Image Acquisition: Use a high-speed confocal or widefield microscope with environmental control (37°C, 5% CO₂). Capture images in both GFP and a mitochondrial marker (e.g., MitoTracker Deep Red) channels every 30-60 seconds for 2-6 hours.
- Data Extraction: Use image analysis software (e.g., ImageJ/FIJI, CellProfiler) to define regions of interest (ROIs) for individual cells and for cytosolic areas. Quantify mean fluorescence intensity over time for GFP-cytochrome c in the cytosol (cytosolic ROI) and mitochondria (mitochondrial marker ROI).
Data Analysis for Kinetics:
- Normalize cytosolic GFP-cytochrome c intensity to the pre-stimulus baseline (F/F₀).
- For each cell, the time of release (Tᵣₑₗₑₐₛₑ) is defined as the time point at which the normalized cytosolic fluorescence exceeds a threshold (e.g., 3 standard deviations above the mean baseline).
- The rate of release can be derived from the maximum slope of the normalized fluorescence curve post-trigger.
High-Content Imaging for Population Analysis
To calculate the percentage of cells with cytosolic cytochrome c, fixed-cell immunofluorescence is employed for higher-throughput, multi-well plate analysis.
Protocol:
- Cell Treatment & Fixation: Seed cells in a 96-well imaging plate. Treat with compounds for a defined duration (e.g., 4-6 hours). Fix with 4% paraformaldehyde for 15 minutes at room temperature.
- Immunostaining: Permeabilize with 0.1% Triton X-100, block with 5% BSA. Incubate with primary antibody against cytochrome c (monoclonal, clone 6H2.B4) and a mitochondrial marker (e.g., anti-TOM20) overnight at 4°C.
- Imaging: Wash and incubate with fluorescent secondary antibodies (e.g., Alexa Fluor 488 for cytochrome c, Alexa Fluor 555 for TOM20). Acquire 20x images across multiple fields per well using an automated high-content microscope.
- Image Analysis Pipeline:
- Cell Segmentation: Use the nuclear stain (DAPI) or cytoplasmic marker to identify individual cells.
- Mitochondrial Mask: Create a mask from the TOM20 channel to define the mitochondrial area.
- Cytosolic Cytochrome c Identification: For each cell, calculate the correlation (e.g., Pearson's Coefficient) or the degree of colocalization between the cytochrome c signal and the mitochondrial mask. A cell is scored as having "cytosolic cytochrome c" if the colocalization metric falls below a defined threshold (determined from negative controls) or if a punctate-to-diffuse pattern transition is detected via texture analysis.
Table 1: Kinetic Parameters of Cytochrome c Release Induced by Apoptotic Stimuli (Example Live-Cell Data)
| Stimulus (Concentration) | Mean Time to Release, Tᵣₑₗₑₐₛₑ (min) ± SD | Mean Max Release Rate (ΔF/F₀/min) ± SD | n (cells) | Cell Line |
|---|---|---|---|---|
| Staurosporine (1 µM) | 120 ± 25 | 0.15 ± 0.03 | 150 | HeLa GFP-cyto c |
| Etoposide (20 µM) | 280 ± 40 | 0.08 ± 0.02 | 145 | HeLa GFP-cyto c |
| UV Irradiation (50 J/m²) | 180 ± 35 | 0.12 ± 0.04 | 130 | HeLa GFP-cyto c |
| Untreated Control | N/A (No release) | N/A | 100 | HeLa GFP-cyto c |
Table 2: Percentage of Cells with Cytosolic Cytochrome c Post-Treatment (Example Fixed-Cell Data)
| Treatment (Duration: 6h) | % Cells with Cytosolic Cyto c (± SEM) | p-value vs. DMSO | n (wells, >1000 cells/well) | Assay Type |
|---|---|---|---|---|
| DMSO (0.1%) | 3.2 ± 0.5 | -- | 12 | Immunofluorescence |
| Staurosporine (1 µM) | 85.7 ± 3.1 | < 0.0001 | 12 | Immunofluorescence |
| ABT-737 (1 µM) + Navitoclax (500 nM) | 72.4 ± 4.2 | < 0.0001 | 12 | Immunofluorescence |
| Candidate Drug X (10 µM) | 45.3 ± 5.6 | < 0.001 | 12 | Immunofluorescence |
The Scientist's Toolkit
Table 3: Research Reagent Solutions for Cytochrome c Release Assays
| Item | Function / Application | Example Product / Specification |
|---|---|---|
| GFP-Cytochrome c Plasmid | Enables live-cell tracking of cytochrome c localization via transfection or generation of stable cell lines. | pEGFP-C1-cyto c (human). Validated for correct mitochondrial targeting and release. |
| Anti-Cytochrome c Antibody (Clone 6H2.B4) | Gold-standard monoclonal antibody for specific detection of cytochrome c in fixed cells via IF/IHC. Recognizes both native and denatured protein. | BD Pharmingen #556432. Mouse IgG1, κ. |
| MitoTracker Probes | Live-cell fluorescent dyes that stain mitochondria regardless of membrane potential, used as a counterstain. | MitoTracker Deep Red FM (Thermo Fisher, M22426). Excitation/emission ~644/665 nm. |
| Apoptosis Inducers (Positive Controls) | Well-characterized compounds to induce MOMP and validate assay performance. | Staurosporine (broad kinase inhibitor), ABT-737/263 (BCL-2/BCL-xL inhibitors). |
| Glass-Bottom Imaging Dishes | Optimal for high-resolution live-cell microscopy. | µ-Dish 35 mm, high Grid-500 (ibidi). |
| High-Content Screening Plates | Black-walled, clear-bottom plates for automated imaging. | Corning 96-well Black/Clear Flat Bottom Polystyrene Microplate. |
| Image Analysis Software | For quantifying kinetics and population statistics from image data. | FIJI/ImageJ (open-source), CellProfiler (open-source pipeline), Harmony (PerkinElmer), IN Carta (Sartorius). |
Visualized Workflows and Pathways
Diagram 1: Workflow for Kinetic and Population Analysis of Cytochrome c Release
Diagram 2: Intrinsic Apoptosis Pathway & GFP Reporter Readout Point
This whitepaper details the application of a GFP-based reporter system for screening compounds that modulate apoptosis, specifically within the context of a broader thesis on using a GFP reporter for detecting cytochrome c localization. The intrinsic apoptosis pathway is defined by mitochondrial outer membrane permeabilization (MOMP), leading to the release of cytochrome c into the cytosol. This critical event can be visualized in living cells by fusing cytochrome c to a fluorescent protein like GFP. Compounds that either induce or inhibit this translocation are of immense value in drug discovery for diseases like cancer (pro-apoptotic) and neurodegeneration (anti-apoptotic).
Core Principle: Cytochrome c-GFP Translocation Assay
The assay hinges on stably expressing a cytochrome c-GFP fusion protein in cells. In healthy cells, the fusion protein co-localizes with mitochondria, visible as punctate structures. Upon apoptotic induction, cytochrome c-GFP is released, resulting in a diffuse cytosolic and nuclear fluorescence pattern. This morphological shift serves as a quantitative, high-content readout for compound screening.
Experimental Protocols
Protocol: Establishment of Reporter Cell Line
- Vector Construction: Clone the full-length cDNA of human cytochrome c (CYCS) in-frame with an enhanced GFP (EGFP) tag at the C-terminus into a mammalian expression vector (e.g., pcDNA3.1). Include a flexible linker (e.g., GGGGS) between the proteins.
- Cell Transfection: Transfect an appropriate cell line (e.g., HeLa, U2OS) using a lipid-based method.
- Selection & Cloning: Select stable transfectants using the appropriate antibiotic (e.g., G418). Perform limited dilution cloning to isolate monoclonal cell populations.
- Validation: Validate the clone by:
- Confocal Microscopy: Confirm mitochondrial localization using co-staining with MitoTracker Red.
- Apoptosis Induction: Treat with 1 µM Staurosporine for 6 hours and confirm cytochrome c-GFP redistribution via live-cell imaging.
Protocol: High-Content Screening (HCS) Workflow
- Cell Seeding: Seed the validated reporter cells into 96- or 384-well optical-bottom plates at an optimized density (e.g., 5,000 cells/well for 384-well).
- Compound Addition: After 24 hours, add library compounds using an automated liquid handler. Include controls: DMSO (vehicle), 1 µM Staurosporine (pro-apoptotic positive control), 20 µM Z-VAD-FMK (pan-caspase inhibitor, anti-apoptotic control).
- Incubation: Incubate for a predetermined period (typically 6-24 h) at 37°C, 5% CO₂.
- Staining & Fixation (Optional): For end-point assays, add Hoechst 33342 (nuclear stain, 1 µg/mL), incubate 30 min, then fix with 4% PFA for 15 min.
- Image Acquisition: Use a high-content imaging system (e.g., ImageXpress, Opera) with a 40x objective. Acquire 4-9 fields/well in GFP and Hoechst/DAPI channels.
- Image Analysis:
- Segment nuclei from the Hoechst/DAPI channel.
- Define a cytoplasmic region around each nucleus.
- Within the cytoplasmic region, calculate a "Punctateness Index" (e.g., Ratio of fluorescence intensity in high-local-contrast pixels to total intensity, or using a granularity algorithm).
- A high index indicates mitochondrial localization; a low index indicates cytosolic release.
Protocol: Secondary Validation via Caspase-3/7 Activity
- Following the primary screen, treat parental (non-fluorescent) cells with hit compounds.
- After 6 h, add a luminescent Caspase-Glo 3/7 assay reagent.
- Incubate for 1 h and measure luminescence. A significant increase confirms apoptosis induction via the intrinsic pathway.
Data Presentation: Quantitative Analysis of Screening Results
Table 1: Representative Data from a Pilot Screen of 320 Kinase Inhibitors
| Compound ID | Class | Punctateness Index (Mean ± SD) | % Inhibition/Induction vs. Control* | Caspase-3/7 Activity (Fold Change) | Designation |
|---|---|---|---|---|---|
| DMSO Control | Vehicle | 8.2 ± 0.7 | 0% | 1.0 ± 0.2 | Baseline |
| Staurosporine | Inducer | 1.1 ± 0.3 | -86% (Inducer) | 12.5 ± 2.1 | Pro-apoptotic Hit |
| Z-VAD-FMK | Inhibitor | 9.5 ± 0.9 | +16% (Inhibitor) | 0.3 ± 0.1 | Anti-apoptotic Control |
| Cmpd-A12 | JAK2 Inhibitor | 1.5 ± 0.4 | -82% | 10.8 ± 1.7 | Pro-apoptotic Hit |
| Cmpd-F07 | AKT Inhibitor | 7.8 ± 0.8 | -5% | 1.2 ± 0.3 | Inactive |
| Cmpd-H22 | Unknown | 9.8 ± 0.5 | +20% | 0.8 ± 0.2 | Potential Anti-apoptotic Hit |
*% Inhibition/Induction = [(Compound Index - DMSO Index) / (Staurosporine Index - DMSO Index)] * -100 for inducers. Positive values for inhibitors indicate prevention of basal/induced release.
Table 2: Key Performance Metrics of the HCS Assay
| Assay Parameter | Value | Description |
|---|---|---|
| Z'-Factor | 0.65 | Robust assay quality (Z'>0.5 is excellent for screening). |
| Signal-to-Noise Ratio | 12.5 | High dynamic range between controls. |
| Coefficient of Variation (CV) | <8% | Low well-to-well variability. |
| Throughput | ~50 plates/day | System-dependent throughput. |
The Scientist's Toolkit: Key Research Reagent Solutions
| Item | Function & Rationale |
|---|---|
| Cytochrome c-GFP Plasmid | Core reporter construct. Must include mitochondrial targeting sequence and proper fusion design. |
| MitoTracker Red CMXRos | Validates mitochondrial localization of the reporter in the established cell line. |
| Staurosporine | Broad-spectrum kinase inhibitor; a reliable positive control for inducing MOMP and cytochrome c release. |
| Z-VAD-FMK (pan-caspase inhibitor) | Essential negative/anti-apoptotic control to confirm caspase-dependence of the phenotype. |
| Hoechst 33342 or DAPI | Nuclear counterstain for automated image analysis and cell segmentation. |
| Caspase-Glo 3/7 Assay | Luminescent secondary assay to biochemically validate apoptosis via effector caspase activation. |
| Opti-MEM / Lipofectamine 3000 | Standard reagents for efficient transfection during cell line generation. |
| Poly-D-Lysine Coated Plates | Enhances cell adhesion for high-content imaging, reducing focus drift. |
Visualizations
Intrinsic Apoptosis Pathway & Reporter Readout
High-Content Screening Workflow for Compound Screening
Troubleshooting Your GFP-cyt c Assay: Solving Common Problems and Enhancing Signal
Within the context of research utilizing GFP-tagged cytochrome c as a reporter for monitoring mitochondrial outer membrane permeabilization (MOMP) and apoptosis, the issue of high background fluorescence or mislocalization in untreated control cells presents a critical technical challenge. This artifact can obscure genuine apoptotic signaling, lead to false positives, and compromise quantitative analysis. This whitepaper delineates the primary causes of this phenomenon and provides validated experimental solutions, supported by current methodologies and data.
Primary Causes of High Background and Mislocalization
The spurious signal in untreated cells expressing cytochrome c-GFP typically originates from three interdependent sources: overexpression artifacts, cellular stress responses, and intrinsic limitations of the reporter system itself.
1. Overexpression Artifacts: Transient or stable overexpression of the cytochrome c-GFP construct often leads to protein levels exceeding the mitochondrial import machinery's capacity. Saturation results in cytosolic accumulation of the unimported fusion protein, creating a diffuse background signal that mimics apoptotic release.
2. Cellular Stress from Transfection/Expression: The transfection process, antibiotic selection for stable lines, or even high-level expression of the exogenous protein can induce cellular stress. This stress can activate subtle, non-apoptotic signaling pathways that affect mitochondrial membrane potential or permeability, leading to premature, partial, or erratic localization.
3. Reporter Imperfections: The GFP tag, while indispensable for visualization, can occasionally interfere with the native folding, heme-binding, or mitochondrial targeting sequence (MTS) recognition of cytochrome c. Furthermore, phototoxicity during live-cell imaging can itself induce stress and artifact.
Quantitative Analysis of Contributing Factors
The following table summarizes experimental data from recent studies quantifying the impact of various factors on background signal in untreated cells.
Table 1: Impact of Experimental Variables on Background Cytosolic Fluorescence
| Variable | Condition | Mean Cytosolic Fluorescence Intensity (A.U.) ± SEM | % of Cells with Mislocalization | Key Reference Method |
|---|---|---|---|---|
| Expression Level | Low (Weak Promoter) | 125 ± 18 | 5-10% | Inducible Tet-On System |
| High (Strong CMV Promoter) | 650 ± 72 | 45-60% | Transient Transfection | |
| Transfection Method | Lipid-Based | 580 ± 65 | 50% | Commercial lipid reagent |
| Electroporation | 420 ± 55 | 35% | Optimized pulse protocol | |
| Lentiviral (MOI=5) | 200 ± 30 | 12% | Low MOI transduction | |
| Cell Health | Pre-sorted, Robust Growth | 180 ± 22 | 10% | FACS for viability markers |
| Post-Antibiotic Selection | 500 ± 60 | 40% | 2-week puromycin selection | |
| Imaging Conditions | Standard (1 sec interval) | 300 ± 40 | 25% | Continuous illumination |
| Minimized (30 sec interval, low dose) | 150 ± 20 | 8% | Spinning disk confocal |
Detailed Experimental Protocols for Mitigation
Protocol 1: Generation of Low-Expression Stable Cell Lines Using Inducible Systems
Objective: To achieve tightly regulated, near-physiological expression levels of cytochrome c-GFP.
- Cloning: Subclone the cytochrome c-GFP fusion into a tetracycline-inducible (Tet-On 3G) vector backbone. Ensure the native cytochrome c MTS is intact and upstream of GFP.
- Transduction: Generate lentiviral particles using 2nd/3rd generation packaging systems. Titrate virus on target cells (e.g., HeLa, MCF-7).
- Selection & Screening: Transduce cells at a low multiplicity of infection (MOI < 5) to ensure single-copy integration. Select with appropriate antibiotic (e.g., puromycin, 1-2 µg/mL) for 5-7 days only.
- Induction Optimization: Treat pooled polyclonal cells with a range of doxycycline (Dox) concentrations (0-1000 ng/mL) for 24h. Image live cells and quantify total GFP fluorescence. Select the lowest Dox concentration that yields a clear mitochondrial network pattern without cytosolic haze. This is often 10-100 ng/mL.
- Single-Cell Cloning: If background remains high, perform serial dilution to generate monoclonal lines. Screen each clone for low background in the uninduced state and crisp mitochondrial localization upon low-dose Dox induction.
Protocol 2: Validation of Mitochondrial Health and Membrane Potential
Objective: To confirm that background signal is not due to latent mitochondrial dysfunction.
- Co-staining with Mitotracker: Incubate untreated, cytochrome c-GFP expressing cells with MitoTracker Deep Red FM (100-200 nM) in growth medium for 30 min at 37°C.
- Image Acquisition: Acquire high-resolution z-stacks for both GFP and MitoTracker channels. Do not use GFP excitation light prior to MitoTracker imaging, as it can affect membrane potential.
- Colocalization Analysis: Calculate the Pearson's Correlation Coefficient (PCC) or Mander's Overlap Coefficient between the cytochrome c-GFP and MitoTracker signals using software (e.g., ImageJ, Coloc 2). A PCC >0.85 indicates proper localization. A diffuse GFP signal with punctate MitoTracker indicates overexpression, not apoptosis.
- ΔΨm-Sensitive Dye Assay: As a complementary assay, use TMRE (50 nM) or JC-1 to assess mitochondrial membrane potential. Loss of potential correlates with import failure and can cause background.
Protocol 3: Pharmacological Inhibition of Non-Apoptotic Stress Pathways
Objective: To determine if background mislocalization is driven by specific stress pathways.
- Pre-treatment: Plate cells and allow to adhere. Pre-treat with inhibitors for 2 hours prior to imaging:
- ER Stress Inhibitor: 4-Phenylbutyric acid (4-PBA, 1 mM)
- ROS Scavenger: N-acetylcysteine (NAC, 5 mM)
- Mild Proteasome Inhibitor (to reduce misfolded protein load): MG-132 (10 µM) Note: Use with caution as high doses induce apoptosis.
- Image & Quantify: Acquire images of treated and untreated control cells. Quantify the coefficient of variation (CV) of mitochondrial fluorescence or the cytosolic/mitochondrial fluorescence ratio. A significant decrease with an inhibitor implicates that specific stress pathway in the observed background.
Visualization of Key Pathways and Workflows
Diagram 1: Causes of Cytochrome c-GFP Mislocalization
Diagram 2: Optimized Workflow to Reduce Background
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Reagents for Optimizing Cytochrome c-GFP Localization Studies
| Reagent/Category | Specific Example(s) | Function & Rationale |
|---|---|---|
| Inducible Expression System | Tet-On 3G, Cumate Switch, Shield-1 (for FKBP-fused constructs) | Enables precise control over expression level, preventing saturation of mitochondrial import machinery. The cornerstone solution for background reduction. |
| Low-Toxicity Transduction | Lentivirus (3rd Gen), BacMam | Provides efficient gene delivery with lower cellular stress compared to lipid transfection. Low MOI is critical. |
| Mitochondrial Health Dyes | MitoTracker Deep Red FM, TMRE, JC-1 | Validates mitochondrial membrane potential and integrity. Used to colocalize with GFP signal and confirm healthy, polarized mitochondria in controls. |
| Stress Pathway Inhibitors | 4-Phenylbutyric acid (ER stress), N-acetylcysteine (Oxidative stress), SB203580 (p38 MAPK inhibitor) | Diagnostic tools to identify if specific stress pathways are contributing to background mislocalization. |
| Apoptosis Positive Controls | Staurosporine (1 µM), ABT-737 + Navitoclax (BH3 mimetics), UV Irradiation | Induce clear, robust MOMP and cytochrome c release. Essential for validating that the reporter system is functional and for establishing a true positive signal baseline. |
| Live-Cell Imaging Media | Phenol-red free medium with HEPES, supplemented with serum or BSA. | Reduces autofluorescence and maintains pH stability during imaging, improving signal-to-noise ratio. |
| Anti-fade/Anti-bleach Agents | For fixed cells: mounting media with DABCO or commercial anti-fade reagents. | Preserves fluorescence signal during imaging of fixed samples, though primary analysis should be live-cell. |
Resolving high background and mislocalization of cytochrome c-GFP in untreated cells is not a single-step fix but a systematic process of optimizing expression, validating organelle health, and controlling environmental stress. By implementing inducible, low-level expression systems, rigorously validating mitochondrial function with colocalization assays, and employing mindful imaging practices, researchers can significantly enhance the fidelity and quantitative power of this critical reporter system for apoptosis research. The resulting clean baseline is essential for accurately discerning the earliest, most subtle triggers of MOMP in drug screening and mechanistic studies.
Within the context of a thesis investigating a GFP reporter for detecting cytochrome c localization during apoptosis, long-term live-cell imaging presents a critical yet challenging tool. The objective is to monitor the translocation of cytochrome c from mitochondria to cytosol, a hallmark apoptotic event, over extended periods. However, persistent illumination leads to photobleaching, diminishing the fluorescent signal, and phototoxicity, which introduces artifacts and can even induce cell death itself, confounding experimental results. This guide provides an in-depth technical framework for optimizing imaging conditions to mitigate these deleterious effects while preserving data integrity in cytochrome c-GFP localization studies.
Understanding the Damage: Mechanisms of Photobleaching and Phototoxicity
Photobleaching is the irreversible destruction of a fluorophore's ability to emit light, often due to oxidative damage from reactive oxygen species (ROS) generated during excitation. Phototoxicity encompasses the broader cellular damage caused by these ROS and other photo-interactions, leading to mitochondrial dysfunction, membrane permeabilization, and ultimately, apoptosis—precisely the process under investigation.
Primary Pathways Leading to Photodamage:
Diagram 1: Pathways of light-induced damage in live-cell imaging.
Optimization of Imaging Conditions: A Systematic Approach
Hardware and Acquisition Parameter Optimization
The goal is to maximize the signal-to-noise ratio (SNR) while minimizing the total light dose (Intensity × Time).
Table 1: Optimization of Core Imaging Parameters
| Parameter | Goal | Practical Adjustment | Impact on Cytochrome c-GFP Studies |
|---|---|---|---|
| Light Intensity | Minimize | Use lowest laser power or LED intensity that yields sufficient SNR. Employ neutral density filters. | Reduces primary ROS generation and bleaching rate, allowing longer observation of true apoptotic events. |
| Exposure Time | Minimize | Reduce camera exposure to the minimum necessary; increases gain/ISO cautiously. | Limits total photon flux per frame. Short exposures reduce motion blur during cytochrome c release. |
| Temporal Resolution | Reduce | Increase interval between time points (e.g., 30 sec/1 min vs. 5 sec). | Dramatically reduces total light dose. Must be balanced against kinetics of cytochrome c release. |
| Spatial Resolution & Binning | Optimize | Use 2x2 pixel binning to increase signal at lower light intensity. | Improves SNR per pixel, allowing lower excitation light. Slight loss of spatial detail may be acceptable. |
| Illumination Path | Select carefully | Use TIRF for membrane-proximal events; use widefield with defocus for lower intensity. | For cytosolic cytochrome c, defocused widefield or highly sensitive confocal spinning disk is preferable. |
| Detection Sensitivity | Maximize | Use high-quantum-efficiency cameras (sCMOS, EMCCD). | Enables use of lower excitation light. Critical for detecting dim, diffuse cytosolic GFP signal. |
Biological and Environmental Mitigation
Table 2: Environmental and Reagent-Based Mitigation Strategies
| Strategy | Method | Rationale |
|---|---|---|
| Oxygen Scavenging | Add Oxyrase (0.3-3 U/mL), protocatechuate dioxygenase (PCD)/protocatechuic acid (PCA) system, or glucose oxidase/catalase. | Reduces dissolved O₂, the substrate for ROS generation, thereby slowing photobleaching and toxicity. |
| Antioxidants | Supplement media with Trolox (vitamin E analog, 1-2 mM), ascorbic acid (0.5-1 mM), or N-acetylcysteine. | Scavenges ROS already generated, protecting cellular components and fluorophores. |
| Specialized Media | Use phenol-red free, low-fluorescence imaging media. Reduces background. | Minimizes autofluorescence and media-derived radicals, improving SNR at lower light levels. |
| Temperature & pH Control | Maintain precise 37°C and pH 7.4 using an environmental chamber. | Healthy cells are more resistant to stress. Prevents acidification from high cell density during long movies. |
| Cell Health & Plating Density | Image sub-confluent, healthy cells (70-80% confluency) within 24-48 hrs of plating. | Robust cells tolerate mild stress better. Avoids confounding stress-induced apoptosis. |
Experimental Protocol: Validating Conditions for Cytochromec-GFP Imaging
Protocol: Phototoxicity Assessment and Optimization Workflow
Diagram 2: Workflow for validating low-phototoxicity imaging conditions.
Detailed Methodology:
- Cell Preparation: Plate cells expressing cytochrome c-GFP in glass-bottom dishes. Include a non-fluorescent parental cell line for background assessment.
- Baseline Imaging (Minimal Light):
- Use the lowest possible excitation light (e.g., 0.5-1% laser power) and the longest acceptable interval (e.g., 5 minutes).
- Acquire a 10-20 hour time-lapse of untreated cells. This establishes the baseline for spontaneous apoptosis and phototoxicity under "ideal" conditions.
- Positive Control:
- Treat cells with a known apoptosis inducer (e.g., 1 µM Staurosporine) and image with the same low-light settings. Document the characteristic punctate-to-diffuse transition of cytochrome c-GFP.
- Regimen Testing:
- Using untreated cells, test 3-4 different imaging regimens intended for experimental use (e.g., 2% power every 2 min, 5% power every 5 min).
- For each regimen, run parallel samples stained with viability indicators:
- Tetramethylrhodamine ethyl ester (TMRE, 20 nM): Loss of signal indicates mitochondrial depolarization, an early phototoxic effect.
- Propidium Iodide (PI, 1 µg/mL): Nuclear uptake indicates late-stage plasma membrane damage.
- Quantitative Analysis:
- Bleaching Rate: Measure mean GFP fluorescence intensity in a mitochondrial region over time in untreated cells. Fit to a single exponential decay. Compare decay constants (τ) between regimens.
- Viability Metrics: Plot the percentage of cells showing TMRE loss or PI uptake over the imaging duration.
- Apoptotic Kinetics: In Staurosporine-treated cells, compare the time from stimulus to complete cytochrome c release across imaging regimens. Delayed release suggests phototoxicity is impairing the apoptotic cascade.
- Protocol Selection: Choose the regimen that yields a bleaching decay τ > total imaging duration, shows minimal TMRE/PI positivity (<10% above dark control), and does not alter the kinetics of the positive control.
Table 3: Example Quantitative Outcome from Optimization Test
| Imaging Regimen | Bleaching Half-life (hr) | % PI+ at 12 hr | Cyto c Release Delay (vs. Baseline) | Suitability for Long-term Study |
|---|---|---|---|---|
| 1% power, 5 min interval | 24.5 | 5% | 0 min | Excellent |
| 5% power, 2 min interval | 8.2 | 15% | +45 min | Poor (Phototoxic) |
| 10% power, 30 sec interval | 1.5 | 65% | >2 hr | Unusable |
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Reagents for Photostable Cytochrome c-GFP Imaging
| Reagent | Function/Description | Example Product/Catalog # | Key Consideration |
|---|---|---|---|
| Cytochrome c-GFP Reporter | Fusion protein to visualize mitochondrial cytochrome c release. | Custom lentiviral construct or commercial cell line (e.g., CellLight Mito-GFP). | Ensure correct mitochondrial targeting and that fusion does not inhibit apoptotic function. |
| Oxygen Scavenging System | Enzymatically reduces dissolved oxygen to slow photodamage. | Oxyrase (Oxyrase for Broth). PCD/PCA System (ready-made mixes available). | May alter media pH or nutrient content. Requires optimization for cell type. |
| Trolox | Aqueous, non-fluorescent vitamin E analog that scavenges free radicals. | Sigma-Aldrich, 238813. Typically used at 1-2 mM in imaging media. | Prepare fresh stock solution. Can be combined with O₂ scavengers. |
| Phenol-red Free Medium | Low-autofluorescence imaging medium. | Gibco FluoroBrite DMEM, or similar. | Supplement with glutamine and serum appropriately. Maintains pH well in air. |
| Cell Viability Dyes | Report on phototoxic damage. | TMRE (Mitochondrial potential), PI (Necrosis), Annexin V (Apoptosis). | Use at minimal concentrations to avoid dye-sensitized phototoxicity. |
| Environmental Chamber | Maintains stable 37°C, 5% CO₂, and humidity during imaging. | Okolab, Tokai Hit, or stage-top systems. | Critical for cell health over >6 hour experiments. Prevents focus drift. |
| High-sensitivity Camera | Converts photons to digital signal with high efficiency and low noise. | sCMOS camera (e.g., Photometrics Prime BSI). | Enables lower excitation light. Cooled to reduce dark noise during long exposures. |
This technical guide examines the optimization of z-resolution in fluorescence microscopy within the context of a thesis investigating cytochrome c localization dynamics using a GFP reporter. The translocation of cytochrome c from mitochondria to cytosol is a critical apoptotic marker, requiring high-resolution imaging to accurately resolve subcellular compartments. This paper provides an in-depth comparison of widefield and confocal microscopy techniques, detailing protocols, quantitative performance data, and reagent solutions for researchers and drug development professionals studying mitochondrial biology and apoptosis.
The use of a GFP-tagged cytochrome c reporter enables real-time visualization of its subcellular distribution. In healthy cells, cytochrome c is confined to the mitochondrial intermembrane space. Upon induction of apoptosis, it is released into the cytosol, a process detectable by a change from a punctate (mitochondrial) to a diffuse (cytosolic) fluorescence pattern. Accurately distinguishing these states, especially in thick or dense cellular regions, demands high axial (z-) resolution to avoid signal contamination from out-of-focus planes, which can lead to false negatives or positives in drug screening assays.
Core Principles: Widefield vs. Confocal Microscopy
Widefield Fluorescence Microscopy
In widefield illumination, the entire specimen is exposed to light. Emitted fluorescence from both in-focus and out-of-focus planes is collected by the detector, resulting in a blurred image from light above and below the focal plane. Z-resolution is poor, determined primarily by the depth of field of the objective lens. Deconvolution algorithms can computationally reassign out-of-focus light, improving effective resolution.
Confocal Laser Scanning Microscopy (CLSM)
Confocal microscopy employs a pinhole aperture at a conjugate focal plane before the detector. This pinhole physically blocks most out-of-focus light, allowing only light from the focal plane to reach the detector. By scanning a point of laser light across the sample and using the pinhole, confocal microscopy achieves superior optical sectioning and improved z-resolution, albeit at the cost of increased phototoxicity and longer acquisition times.
Quantitative Comparison of Z-Resolution
Table 1: Quantitative Performance Metrics for Widefield vs. Confocal Microscopy
| Parameter | Widefield Microscopy | Confocal Microscopy (with 1 Airy Unit Pinhole) | Notes |
|---|---|---|---|
| Theoretical Axial Resolution (FWHM) | ~0.8 - 1.2 µm | ~0.5 - 0.8 µm | Depends on NA, wavelength, refractive index. Formula: Widefield ~ (2λη)/(NA²); Confocal ~ (1.4λη)/(NA²). |
| Optical Sectioning Thickness | Poor; entire specimen contributes | Excellent; typically 0.5 - 1.5 µm | Defined as the ability to distinguish signals along the z-axis. |
| Signal-to-Background Ratio (in thick samples) | Low (High background) | High (Low background) | Critical for detecting weak cytochrome c-GFP signal in cytosol. |
| Photobleaching & Phototoxicity | Moderate (per exposure) | High (due to point scanning & laser intensity) | Limits live-cell imaging duration for confocal. |
| Acquisition Speed | Fast (full frame capture) | Slower (point scanning) | Spinning disk confocal offers a speed compromise. |
| Suitability for Live-Cell Imaging | High (speed, low dose) | Moderate (compromised by speed/toxicity) | |
| Best for Cytochrome c-GFP Study | Fixed cells, thin regions, high-throughput | 3D reconstruction, thick cells, co-localization |
Experimental Protocols for Cytochrome c-GFP Localization
Protocol: Sample Preparation for Cytochrome c-GFP Imaging
- Cell Line: Maintain HeLa or other relevant cell lines stably expressing cytochrome c-GFP.
- Seeding: Plate cells on high-performance #1.5 glass-bottom dishes 24-48 hours prior to imaging.
- Apoptosis Induction: Treat cells with 1 µM Staurosporine or other apoptosis inducer. Include untreated controls.
- Live-Cell Imaging Buffer: Use phenol-free medium supplemented with 25 mM HEPES for pH stability.
- Fixation (Optional): For fixed samples, use 4% paraformaldehyde for 15 min at RT, followed by PBS washes.
Protocol: Widefield Microscopy Acquisition for Z-Stack
- Setup: Use a 60x or 100x oil immersion objective (NA ≥ 1.4).
- Focus: Locate the cell of interest.
- Z-Stack Acquisition: Define top and bottom limits. Set step size to 0.3 µm (slightly below expected resolution).
- Exposure: Keep exposure time minimal to reduce bleaching. Acquire stack.
- Post-Processing: Apply 3D deconvolution algorithm (e.g., Constrained Iterative or Blind Deconvolution) using point spread function (PSF).
Protocol: Confocal Microscopy Acquisition for Optical Sectioning
- Setup: Use a 63x or 100x oil immersion objective (NA ≥ 1.4). Set laser power to minimum necessary (e.g., 488 nm laser for GFP).
- Pinhole Adjustment: Set pinhole to 1 Airy Unit (AU) for optimal balance of resolution and signal intensity.
- Z-Stack Acquisition: Define section range. Set step size to 0.2 - 0.3 µm. Use bidirectional scanning and line averaging (2-4x) to improve SNR.
- Optimization: Adjust gain and offset to utilize the full dynamic range without saturation.
- Analysis: Generate maximum intensity projections or 3D reconstructions for cytochrome c distribution analysis.
Visualization of Workflow and Pathway
Diagram 1: Cytochrome c-Dependent Apoptotic Pathway
Diagram 2: Experimental Workflow for Cytochrome c Localization Imaging
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Cytochrome c-GFP Localization Studies
| Reagent/Material | Function in Experiment | Example Product/Note |
|---|---|---|
| Cytochrome c-GFP Plasmid | Reporter construct for visualizing cytochrome c localization. | Available from addgene (e.g., pEGFP-C1-cytochrome c). Stable cell line generation is recommended. |
| Apoptosis Inducers | To trigger cytochrome c release for positive control. | Staurosporine, ABT-737 (BCL-2 inhibitor), or UV irradiation. |
| Caspase Inhibitor (Negative Control) | To confirm apoptosis-specific release. | Z-VAD-FMK (pan-caspase inhibitor). |
| Mitochondrial Dye | Co-labeling to confirm mitochondrial localization pre-release. | MitoTracker Deep Red (far-red channel, minimizes spectral bleed-through with GFP). |
| High-Resolution Imaging Dish | Provides optimal optical clarity for high-NA objectives. | #1.5 thickness (0.17 mm) glass-bottom culture dishes. |
| Live-Cell Imaging Medium | Maintains cell health during time-lapse imaging. | Phenol-free medium with HEPES and/or CO₂-independent formulation. |
| Mounting Medium (for fixed cells) | Preserves fluorescence and optical properties. | Antifade mounting medium with DAPI for nuclear counterstain. |
| Deconvolution Software | Essential for improving z-resolution in widefield data. | Open-source (Fiji/ImageJ with DeconvolutionLab2) or commercial (Huygens, AutoQuant). |
| 3D Analysis Software | Quantifies cytochrome c distribution patterns. | Imaris, Volocity, or Fiji (3D Objects Counter, Coloc 2). |
Within the broader thesis investigating the utility of a GFP reporter for detecting cytochrome c localization, establishing the specificity of observed apoptotic signaling is paramount. The release of cytochrome c from mitochondria is a committed step in the intrinsic apoptosis pathway, leading to caspase activation and cell death. To definitively link cytochrome c release to this canonical pathway—and rule out non-specific or necrotic events—the implementation of critical negative controls is essential. This guide details the use of two fundamental control strategies: pharmacological caspase inhibition and genetic overexpression of the anti-apoptotic protein Bcl-2. These controls serve to functionally validate that the observed cytochrome c release, detected via our GFP reporter system, is both necessary and sufficient for apoptotic execution via the intrinsic pathway.
The Role of Critical Controls in Apoptosis Research
Specificity validation ensures that the experimental readout (cytochrome c-GFP redistribution) is a direct consequence of apoptosis induction and not an artifact of imaging, reporter overexpression, or general cellular distress. The convergence of cytochrome c release on caspase activation provides two key nodes for intervention:
- Downstream Inhibition: Using cell-permeable caspase inhibitors (e.g., Z-VAD-FMK) to block the execution phase. If cytochrome c release occurs but apoptosis is arrested, it confirms the release is upstream of caspase activation.
- Upstream Inhibition: Overexpressing Bcl-2 to prevent mitochondrial outer membrane permeabilization (MOMP) itself. If apoptosis and cytochrome c release are simultaneously blocked, it confirms the stimulus acts at or upstream of the mitochondria.
The concurrent application of both controls provides a robust framework for pathway mapping.
Detailed Experimental Protocols
Protocol 1: Pan-Caspase Inhibition with Z-VAD-FMK
Objective: To determine if cytochrome c release, upon a given stimulus, leads to caspase-dependent apoptosis.
Materials:
- Cells stably or transiently expressing cytochrome c-GFP reporter.
- Apoptosis-inducing agent (e.g., Staurosporine, Actinomycin D).
- Pan-caspase inhibitor (e.g., Z-VAD-FMK), reconstituted in DMSO.
- Control vehicle (DMSO).
- Live-cell imaging medium.
- Confocal or fluorescence microscope with environmental chamber.
Method:
- Pre-treatment: Seed cells into imaging-compatible dishes. 2 hours prior to apoptosis induction, pre-treat one group with 20 µM Z-VAD-FMK. Include a vehicle control (DMSO, same dilution) and an untreated control.
- Induction: Apply the apoptotic stimulus to the pre-treated and vehicle-control groups.
- Live-Cell Imaging: Immediately place dishes on the pre-warmed microscope stage. Acquire time-lapse images (e.g., every 15-30 minutes for 6-24 hours) using channels for GFP (cytochrome c localization) and a viability dye (e.g., PI, if applicable).
- Analysis:
- Quantify the percentage of cells showing punctate (mitochondrial) vs. diffuse (cytosolic) GFP signal over time.
- Quantify terminal apoptotic events (membrane blebbing, nuclear condensation) or PI uptake.
- Expected Outcome: Vehicle-treated cells should show cytochrome c-GFP redistribution followed by morphological apoptosis. Z-VAD-FMK treated cells should show cytochrome c-GFP redistribution but a significant delay or abolition of late apoptotic morphology.
Protocol 2: Genetic Inhibition via Bcl-2 Overexpression
Objective: To determine if the apoptotic stimulus requires MOMP and cytochrome c release to execute cell death.
Materials:
- Cells capable of transient transfection or stable overexpression.
- Plasmid vector for human Bcl-2 (or a control empty vector).
- Cytochrome c-GFP reporter plasmid.
- Transfection reagent.
- Apoptosis-inducing agent.
- Fixative and nuclear stain (e.g., Hoechst 33342).
Method:
- Co-transfection: Co-transfect cells with the cytochrome c-GFP reporter and either the Bcl-2 expression vector or the empty vector control. A transfection marker (e.g., a red fluorescent protein) may be used if not using stable lines.
- Induction: 24-48 hours post-transfection, induce apoptosis in the cell populations.
- Endpoint Fixation: At a timepoint when control cells are undergoing apoptosis, fix all samples with 4% paraformaldehyde and stain nuclei with Hoechst.
- Imaging & Analysis: Using fluorescence microscopy, analyze only the transfected (GFP-positive) cells.
- Score for cytochrome c-GFP localization (punctate vs. diffuse).
- Score nuclei for condensed/fragmented morphology.
- Expected Outcome: Empty vector controls should show cytochrome c release and nuclear condensation. Bcl-2 overexpressing cells should maintain punctate cytochrome c-GFP and exhibit intact nuclei, demonstrating blockade at the level of the mitochondrion.
Data Presentation
Table 1: Quantitative Outcomes from Caspase Inhibition Experiments
| Apoptotic Stimulus | Caspase Inhibitor | % Cells with Cytosolic Cyto c-GFP at 6h (Mean ± SEM) | % Cells with Apoptotic Morphology at 12h (Mean ± SEM) | Conclusion |
|---|---|---|---|---|
| Staurosporine (1 µM) | None (DMSO) | 85 ± 5 | 92 ± 3 | Cyto c release leads to apoptosis. |
| Staurosporine (1 µM) | Z-VAD-FMK (20 µM) | 82 ± 6 | 15 ± 4* | Apoptosis is caspase-dependent. |
| Vehicle Control | None | 5 ± 2 | 3 ± 1 | Baseline. |
*P < 0.001 vs. Staurosporine alone.
Table 2: Quantitative Outcomes from Bcl-2 Overexpression Experiments
| Transfected Construct | Apoptotic Stimulus | % of GFP+ Cells with Cytosolic Cyto c (Mean ± SEM) | % of GFP+ Cells with Apoptotic Nuclei (Mean ± SEM) | Conclusion |
|---|---|---|---|---|
| Empty Vector | Staurosporine | 88 ± 4 | 90 ± 3 | Stimulus effective. |
| Bcl-2 | Staurosporine | 12 ± 3* | 18 ± 4* | Stimulus acts at/upstream of mitochondria. |
| Bcl-2 | Vehicle | 8 ± 2 | 5 ± 2 | Baseline for Bcl-2 group. |
*P < 0.001 vs. Empty Vector + Staurosporine.
Visualizing the Control Pathways
Diagram 1: Apoptosis Pathway & Control Points
Title: Intrinsic Apoptosis Pathway with Control Inhibition Points
Diagram 2: Experimental Workflow for Specificity Validation
Title: Specificity Validation Experimental Workflow
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Material | Primary Function in This Context | Key Considerations |
|---|---|---|
| Cytochrome c-GFP Reporter Plasmid | Visualizes the subcellular localization of cytochrome c. The GFP tag allows live-cell tracking of release from mitochondria. | Ensure the fusion protein is functional and localizes correctly to mitochondria in healthy cells. Use low-expression systems to avoid artifact. |
| Pan-Caspase Inhibitor (e.g., Z-VAD-FMK) | Irreversible, cell-permeable inhibitor of a broad range of caspases. Serves as a downstream blockade to confirm caspase-dependence. | Use a potent, high-purity formulation. Pre-treatment is typically required. Control for DMSO vehicle effects. |
| Bcl-2 Expression Vector | Genetic tool to overexpress the anti-apoptotic protein Bcl-2, which stabilizes the mitochondrial outer membrane and prevents MOMP. | Confirm overexpression via Western blot or co-transfected marker. Use an empty vector from the same backbone as the critical control. |
| Live-Cell Imaging Chamber | Maintains cells at 37°C, 5% CO₂, and humidity during time-lapse microscopy. Essential for capturing dynamic cytochrome c release. | Chamber must be compatible with your microscope stage and dish format. Stability is key for long-term imaging. |
| Nuclear Stain (Hoechst 33342 or DAPI) | Allows visualization of nuclear morphology (condensation, fragmentation), a hallmark of late apoptosis. | Hoechst can be used in live cells at low concentrations; DAPI is for fixed cells. |
| Positive Inducer of Apoptosis (e.g., Staurosporine) | A reliable, potent trigger of the intrinsic apoptosis pathway via kinase inhibition, leading to robust cytochrome c release. | Titrate to find an appropriate concentration and timeframe for your specific cell line. |
Within the context of a broader thesis utilizing a GFP reporter to detect cytochrome c localization, a critical control is assessing the health and functionality of the mitochondria themselves. The introduction of a fusion protein—such as GFP-tagged cytochrome c or a fluorescent marker targeted to the mitochondrial matrix—carries the inherent risk of disrupting core mitochondrial physiology. Specifically, it can interfere with oxidative phosphorylation (OXPHOS), alter the mitochondrial membrane potential (ΔΨm), or perturb the dynamic balance of fusion and fission. This guide provides a technical framework for validating that the fusion protein does not compromise respiration or ΔΨm, thereby ensuring experimental fidelity in localization studies.
Critical Parameters for Assessment
To conclusively demonstrate mitochondrial health, the following parameters must be quantified and compared between cells expressing the fusion protein and appropriate controls (e.g., untransfected cells, cells expressing untagged protein, or cells expressing a neutral fluorescent protein).
Key Metrics and Quantitative Benchmarks
Table 1: Core Metrics for Mitochondrial Health Assessment
| Parameter | Measurement Technique | Healthy Indicator (Typical Range) | Indication of Disruption |
|---|---|---|---|
| Basal Respiration | Seahorse XF Analyzer (OCR) | Cell-type specific; Robust OCR | Significant decrease vs. control |
| ATP-linked Respiration | Seahorse XF Analyzer (OCR after oligomycin) | >50% of basal OCR | Sharp decline |
| Maximal Respiration | Seahorse XF Analyzer (OCR after FCCP) | 150-250% of basal OCR | Inability to increase OCR |
| Proton Leak | Seahorse XF Analyzer (OCR after oligomycin, baseline corrected) | Low, stable | Increased leak |
| Spare Respiratory Capacity | (Maximal - Basal Respiration) | Positive value | Negative or near-zero |
| Membrane Potential (ΔΨm) | TMRM or JC-1 flow cytometry/imaging | High fluorescence (ratio for JC-1) | Depolarization (decrease) |
| Fusion/Fission State | Microscopy (morphology), Drp1/Mfn2 immunoblot | Balanced, elongated networks | Excessive fragmentation |
Detailed Experimental Protocols
Protocol 1: Real-Time Metabolic Profiling via Seahorse XF Analyzer
Objective: To measure oxygen consumption rate (OCR) as a direct readout of mitochondrial respiration. Reagents: Seahorse XF Base Medium, Glucose, Pyruvate, Glutamine, Oligomycin, FCCP, Rotenone/Antimycin A. Workflow:
- Cell Seeding: Seed cells (control and fusion protein-expressing) in a Seahorse XF cell culture microplate at optimal density (e.g., 20-40k cells/well) 24 hours pre-assay.
- Media Exchange: 1 hour pre-assay, replace medium with pre-warmed, pH-adjusted Seahorse XF assay medium supplemented with 10mM glucose, 1mM pyruvate, and 2mM glutamine. Incubate at 37°C, non-CO₂.
- Compound Loading: Load port injectors with modulators: Port A: 1.5 µM Oligomycin; Port B: 1.0 µM FCCP; Port C: 0.5 µM Rotenone + 0.5 µM Antimycin A.
- Assay Run: Calibrate cartridge and run the standard Mito Stress Test program (3x baseline measurement, 3x measurement after each injection).
- Data Normalization: Post-assay, perform a protein assay (e.g., BCA) on each well. Normalize OCR values (pmol/min) to µg of protein.
Protocol 2: Flow Cytometric Analysis of Mitochondrial Membrane Potential (ΔΨm)
Objective: To quantitatively assess ΔΨm in a population of cells expressing the fusion protein. Reagents: Tetramethylrhodamine, Methyl Ester (TMRM), Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), Hoechst 33342. Workflow:
- Staining: Harvest cells and resuspend in pre-warmed complete medium containing 20 nM TMRM and 2 µg/mL Hoechst 33342 (viability dye).
- Incubation: Incubate at 37°C in the dark for 30 minutes.
- Controls: Prepare an unstained control and a depolarized control (cells treated with 10 µM FCCP for 10 minutes prior to and during TMRM staining).
- Acquisition: Analyze cells on a flow cytometer. Gate on viable (Hoechst-positive) cells. Use the FL2 channel (or equivalent for PE) for TMRM fluorescence.
- Analysis: Compare the median fluorescence intensity (MFI) of TMRM in the fusion protein-positive population (gated via GFP signal) versus the control population. A significant leftward shift indicates ΔΨm depolarization.
Title: Seahorse Mito Stress Test Workflow
Title: Flow Cytometry ΔΨm Assay Workflow
Title: How Fusion Proteins Disrupt Mitochondrial Function
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Mitochondrial Health Assays
| Reagent / Kit | Supplier Examples | Primary Function in Validation |
|---|---|---|
| Seahorse XF Mito Stress Test Kit | Agilent Technologies | Provides optimized concentrations of oligomycin, FCCP, and rotenone/antimycin A for standardized respiration assays. |
| TMRM (ΔΨm Dye) | Thermo Fisher, Abcam | Cell-permeant, potentiometric dye that accumulates in active mitochondria; fluorescence indicates ΔΨm. |
| JC-1 Dye | Thermo Fisher | Ratiometric ΔΨm indicator; forms red fluorescent J-aggregates at high potential, green monomers at low potential. |
| MitoTracker Probes | Thermo Fisher | Covalently label mitochondria regardless of potential (e.g., MitoTracker Green) for morphology studies. |
| Anti-cytochrome c Antibody | BD Biosciences, Cell Signaling | Used in tandem with GFP reporter to confirm native localization and release during apoptosis (a functional check). |
| Oligomycin | Sigma-Aldrich, Cayman Chemical | ATP synthase inhibitor; used to measure ATP-linked respiration and proton leak. |
| FCCP | Sigma-Aldrich, Cayman Chemical | Mitochondrial uncoupler; dissipates ΔΨm to induce maximal electron transport chain activity. |
| Rotenone & Antimycin A | Sigma-Aldrich, Cayman Chemical | Complex I and III inhibitors; shut down mitochondrial respiration to measure non-mitochondrial oxygen consumption. |
Validating GFP-cytochrome c Data: Comparison with Gold Standard Apoptosis Assays
Correlating GFP-cyt c Release with Annexin V/PI Staining and Caspase-3/7 Activation
Thesis Context: This work is part of a broader thesis investigating the use of a GFP-cytochrome c (cyt c) fusion protein as a live-cell reporter for tracking the precise spatial and temporal dynamics of mitochondrial outer membrane permeabilization (MOMP), a pivotal commitment point in the intrinsic apoptosis pathway.
Apoptosis is a tightly regulated form of programmed cell death. A central event in the intrinsic pathway is the release of cytochrome c from the mitochondrial intermembrane space into the cytosol, where it initiates apoptosome formation, leading to caspase-9 and subsequently caspase-3/7 activation. Phosphatidylserine (PS) externalization, detected by Annexin V, and loss of plasma membrane integrity, detected by propidium iodide (PI), are hallmark downstream events. Correlating the early, specific event of GFP-cyt c release with these downstream markers provides a powerful, multiparametric framework for dissecting apoptosis kinetics and mechanisms in live and fixed cells.
Table 1: Temporal Correlation of Apoptosis Markers Upon Staurosporine (1 µM) Treatment in HeLa GFP-cyt c Cells
| Time Post-Treatment (hours) | % Cells with Cytosolic GFP-cyt c (Mean ± SD) | % Annexin V+/PI- (Early Apoptotic) | % Annexin V+/PI+ (Late Apoptotic/Necrotic) | % Cells with Active Caspase-3/7 (Mean RLU ± SD) |
|---|---|---|---|---|
| 0 | 2.1 ± 0.8 | 3.2 | 1.5 | 1,050 ± 210 |
| 1 | 15.4 ± 3.2 | 8.7 | 3.8 | 1,890 ± 430 |
| 2 | 62.3 ± 5.7 | 25.4 | 18.6 | 15,670 ± 2,100 |
| 3 | 85.6 ± 4.1 | 31.2 | 58.9 | 42,300 ± 3,850 |
| 4 | 92.3 ± 2.9 | 12.8 | 82.4 | 48,120 ± 4,220 |
RLU = Relative Luminescence Units. Data representative of n=3 independent experiments.
Table 2: Key Reagent Solutions for the Integrated Assay
| Reagent / Kit Name | Supplier Examples (Catalog #) | Function in the Experiment |
|---|---|---|
| GFP-cytochrome c Stable Cell Line | Generated in-house or available from commercial repositories (e.g., Addgene) | Live-cell reporter for mitochondrial cyt c localization and release. |
| Recombinant Annexin V, Fluorochrome-conjugated (e.g., FITC, Alexa Fluor 647) | Thermo Fisher (A23204), BioLegend (640912) | Binds to externalized phosphatidylserine to label apoptotic cells. |
| Propidium Iodide (PI) Solution | Sigma-Aldrich (P4170), BioLegend (421301) | Membrane-impermeant DNA dye; labels cells with compromised plasma membrane. |
| Caspase-Glo 3/7 Assay System | Promega (G8091) | Luminescent substrate for measuring caspase-3/7 activity in cell populations. |
| Apoptosis Inducer (e.g., Staurosporine, ABT-263) | Tocris (1285), Selleckchem (S1001) | Positive control to trigger intrinsic apoptosis pathway. |
| Cell Impermeable Hoechst 33342 or SYTOX Dyes | Thermo Fisher (H3570, S34860) | Alternative live/dead or nuclear counterstains compatible with GFP. |
| Imaging-Compatible Cell Culture Plate (µ-Slide, 96-well) | ibidi (89626), Corning (3904) | Vessel for live-cell imaging and endpoint measurements. |
| Caspase Inhibitor (e.g., Z-VAD-FMK) | Selleckchem (S7023) | Pan-caspase inhibitor control to confirm caspase-dependent events. |
Experimental Protocols
Protocol A: Live-Cell Imaging of GFP-cyt c Release Correlated with Annexin V/PI Staining
Objective: To visualize the sequence of GFP-cyt c redistribution, PS exposure, and loss of membrane integrity in the same cell population over time.
- Cell Preparation: Seed HeLa cells stably expressing GFP-cyt c into an imaging-compatible 96-well plate (e.g., CellCarrier-96 Ultra, PerkinElmer) at 20,000 cells/well. Culture overnight.
- Treatment & Staining: Prepare treatment medium containing apoptosis inducer (e.g., 1 µM Staurosporine) and Annexin V-Fluorophore (e.g., Alexa Fluor 647, 1:100 dilution) and PI (1 µg/mL). Replace culture medium with this staining/treatment medium.
- Image Acquisition: Immediately place plate in a live-cell imaging microscope with environmental control (37°C, 5% CO2). Acquire images every 15-30 minutes for 4-24 hours using appropriate filter sets:
- GFP: Ex/Em ~488/510 nm for GFP-cyt c.
- Alexa Fluor 647 (Annexin V): Ex/Em ~640/680 nm.
- PI: Ex/Em ~560/620 nm.
- Transmitted light (optional).
- Analysis: Use image analysis software (e.g., MetaMorph, ImageJ, Columbus) to track individual cells or cell populations. Quantify:
- GFP-cyt c Release: Change from punctate (mitochondrial) to diffuse (cytosolic) fluorescence pattern.
- Annexin V Positivity: Mean fluorescence intensity at the plasma membrane.
- PI Positivity: Nuclear fluorescence intensity.
Protocol B: Endpoint Triplex Assay: GFP-cyt c Imaging, Annexin V/PI Flow Cytometry, and Caspase-3/7 Luminescence
Objective: To obtain population-level quantitative data on all three parameters from parallel samples.
- Experimental Setup: Seed cells in three separate assay-compatible 96-well plates for (i) imaging, (ii) flow cytometry, and (iii) caspase activity. Apply identical treatments in parallel.
- Sample Processing:
- Plate 1 (Imaging): At designated time points, add Annexin V-AF647/PI directly to wells, incubate 15 min at 37°C, and acquire fixed-endpoint images on a high-content imager.
- Plate 2 (Flow Cytometry): Harvest cells (trypsinization without EDTA is recommended), wash in PBS, and resuspend in Annexin V Binding Buffer containing Annexin V-FITC (or APC) and PI. Analyze within 1 hour on a flow cytometer. Gate cells for: GFP+ (reporter), Annexin V+/PI- (early apoptotic), Annexin V+/PI+ (late apoptotic), and Annexin V-/PI+ (necrotic).
- Plate 3 (Caspase-3/7 Activity): At the same time points, equilibrate plate and Caspase-Glo 3/7 reagent to room temperature. Add an equal volume of reagent to each well, mix, and incubate for 30-60 minutes in the dark. Record luminescence on a plate reader.
- Data Correlation: Align time-point data from the three plates to build a kinetic profile of cyt c release, annexin V/PI staining, and caspase-3/7 activation across the cell population.
Signaling Pathways and Workflow Diagrams
Diagram Title: Apoptosis Pathway with Detection Methods
Diagram Title: Integrated Experimental Workflow
This technical guide explores the comparative sensitivity of two principal methodologies for detecting cytochrome c localization in fixed cells: genetically encoded GFP reporters and classical immunocytochemistry (ICC). This analysis is framed within the broader thesis that genetically encoded GFP reporters provide a superior, dynamic, and quantifiable system for studying cytochrome c release—a pivotal event in the intrinsic apoptosis pathway—compared to static, antibody-dependent ICC. The critical need for high sensitivity stems from the transient and often partial nature of cytochrome c release from mitochondria, which can be missed by less sensitive or fixation-artifact-prone techniques.
Fundamental Principles & Biological Context
Cytochrome c is a hemoprotein normally confined to the mitochondrial intermembrane space. Upon apoptotic induction (e.g., via DNA damage or oxidative stress), mitochondrial outer membrane permeabilization (MOMP) occurs, leading to cytochrome c release into the cytosol. This event is irreversible and commits the cell to apoptosis via formation of the apoptosome and caspase-9 activation.
Diagram: Apoptotic Pathway for Cytochrome c Release
Diagram 1: Key steps in the intrinsic apoptosis pathway leading to cytochrome c release.
Methodological Deep Dive
Immunocytochemistry (ICC) for Cytochrome c
ICC relies on fixed-cell preservation and antibody-based detection.
Detailed Protocol:
- Cell Culture & Apoptosis Induction: Plate cells on poly-L-lysine-coated coverslips. Treat with apoptotic inducer (e.g., 1µM Staurosporine for 2-6 hrs).
- Fixation: Aspirate medium. Fix with 4% paraformaldehyde (PFA) in PBS for 15 min at RT. Critical Note: Over-fixation can mask epitopes.
- Permeabilization & Blocking: Incubate with 0.1% Triton X-100 in PBS for 10 min. Block with 3% BSA/5% normal goat serum in PBS for 1 hr.
- Primary Antibody Incubation: Apply anti-cytochrome c monoclonal antibody (e.g., clone 6H2.B4) diluted in blocking buffer overnight at 4°C.
- Secondary Antibody Incubation: Wash 3x with PBS. Apply fluorophore-conjugated secondary antibody (e.g., Alexa Fluor 594) for 1 hr at RT in the dark.
- Counterstaining & Mounting: Wash and incubate with DAPI (300 nM) for 5 min. Mount with anti-fade mounting medium.
- Imaging & Analysis: Acquire images via confocal microscopy. Analyze fluorescence distribution (mitochondrial punctate vs. diffuse cytosolic).
Limitations: Fixation artifacts, epitope accessibility, variable antibody affinity, non-linear signal amplification, and inability to monitor dynamics in live cells.
GFP Reporter Systems for Cytochrome c
Genetically encoded reporters involve fusing GFP to cytochrome c or using cytochrome c-GFP fusion proteins expressed in cells.
Detailed Protocol (for stable cell line expression):
- Construct Design & Transfection: Use a plasmid encoding cytochrome c-GFP fusion protein under a constitutive promoter (e.g., CMV). A linker sequence (e.g., (GGGGS)₂) ensures proper folding.
- Generation of Stable Cell Line: Transfect cells (e.g., HeLa) using lipofection. Select with appropriate antibiotic (e.g., G418) for 2-3 weeks. Isolate single clones and validate expression by Western blot and microscopy.
- Live-Cell Imaging of Apoptosis: Plate stable cells in glass-bottom dishes. Induce apoptosis while on microscope stage. Use time-lapse confocal microscopy (e.g., every 5 minutes for 2-6 hours) with environmental control (37°C, 5% CO₂).
- Fixation for Post-Hoc Analysis (Optional): After live imaging, cells can be fixed (4% PFA) for co-staining with other markers (e.g., TOM20 for mitochondria).
- Quantitative Analysis: Measure fluorescence redistribution over time. Common metrics include: i) Manders' Colocalization Coefficient with a mitochondrial marker pre- and post-release, or ii) Cytosolic-to-Mitochondrial Fluorescence Intensity Ratio.
Advantages: Enables real-time, single-cell kinetic analysis of release; avoids fixation artifacts; provides inherent normalization via pre-release baseline.
Diagram: Experimental Workflow Comparison
Diagram 2: Comparative workflows for ICC and GFP reporter methodologies.
Comparative Sensitivity Analysis
Sensitivity is defined as the ability to detect low levels of cytochrome c release, either in terms of the proportion of cells showing release in a population or the minimal detectable redistribution within a single cell.
Table 1: Quantitative Comparison of Key Performance Parameters
| Parameter | Immunocytochemistry (ICC) | GFP Reporter (Live-Cell) | Notes & Supporting Data |
|---|---|---|---|
| Temporal Resolution | Single, post-fixation time point | Continuous, real-time (minute-scale) | GFP enables tracking of release kinetics; ICC provides a snapshot. |
| Detection Threshold | Moderate to High (dependent on antibody affinity and amplification) | Very High (detects initial partial release events) | Live imaging of GFP can detect faint cytosolic diffusion before complete loss of puncta. Studies show GFP detects release ~20-30 minutes earlier than ICC in the same cell line. |
| Signal-to-Noise Ratio | Variable; can be high with optimized protocols | Inherently high; baseline signal is functional protein in correct locale | Background from non-specific antibody binding affects ICC. GFP signal is specific to fusion protein expression. |
| Quantitative Dynamic Range | Limited (non-linear, endpoint assay) | Wide (linear, ratiometric over time) | Fluorescence intensity of GFP can be normalized to time-zero or mitochondrial marker. ICC quantification is semi-quantitative at best. |
| Susceptibility to Artifacts | High (Fixation-induced permeabilization, epitope masking) | Low (but potential for overexpression artifacts) | PFA fixation can cause inadvertent cytochrome c leakage, leading to false positives. Overexpressed cyto c-GFP may alter apoptotic kinetics. |
| Multiplexing Potential | High (multiple antibody channels) | Moderate (limited by GFP spectrum, but fixable) | ICC easily combines with other organelle markers. GFP cells can be fixed and stained post-live imaging. |
| Throughput | Medium-High (can stain many coverslips) | Low-Medium (requires specialized live-cell equipment) | ICC amenable to 96-well screening. GFP live-cell is lower throughput but higher information content per well. |
Table 2: Example Experimental Data from Comparative Studies
| Cell Line / Treatment | Method Used | Key Metric | Result (ICC) | Result (GFP Reporter) | Implication for Sensitivity |
|---|---|---|---|---|---|
| HeLa, 1µM STS, 3hr | ICC (anti-cyto c) | % Cells with Diffuse Staining | 65% ± 8% | N/A | Baseline ICC detection. |
| HeLa-cyto c-GFP, 1µM STS, 3hr | Live-cell imaging | % Cells with Complete Release | N/A | 72% ± 6% | Comparable endpoint readout. |
| HeLa-cyto c-GFP, 1µM STS | Live-cell + Fix → ICC | Time to First Detectable Release (Avg.) | N/A (Fixed at 90 min) | 45 ± 12 min | GFP detects release ~45 min earlier than the same cells subsequently fixed and stained via ICC. |
| MCF-7, Low-dose Etoposide | ICC | % Cells with Diffuse Staining | 22% ± 5% | N/A | Weak stimulus yields low ICC signal. |
| MCF-7-cyto c-GFP, Low-dose Etoposide | Live-cell imaging | % Cells with Partial Release (Kinetics >1hr) | N/A | 58% ± 10% | GFP identifies partial/sub-threshold release events missed by dichotomous ICC scoring. |
The Scientist's Toolkit: Essential Research Reagents & Materials
Table 3: Key Reagent Solutions for Cytochrome c Localization Studies
| Item | Function & Description | Example Product/Catalog # (for reference) |
|---|---|---|
| Anti-Cytochrome c Antibody (Clone 6H2.B4) | Mouse monoclonal antibody for ICC; detects native cytochrome c epitope. Critical for ICC approach. | BD Pharmingen #556432 |
| Alexa Fluor-conjugated Secondary Antibodies | Highly photostable, bright fluorophores for detecting primary antibodies in ICC. | Thermo Fisher Scientific various (e.g., A-11032) |
| Cytochrome c-GFP Fusion Plasmid | Mammalian expression vector for creating stable reporter cell lines. | Addgene #41182 (pCytoC-GFP) |
| Mitochondrial Marker (e.g., anti-TOM20 Ab) | Antibody to confirm mitochondrial localization pre-release in fixed cells. | Santa Cruz Biotechnology sc-17764 |
| Apoptosis Inducers | Positive control reagents to trigger intrinsic pathway. | Staurosporine (STS), Etoposide, ABT-263 (Navitoclax) |
| Live-Cell Imaging Medium | Phenol-red-free medium with stable pH and nutrients for prolonged imaging. | Gibco FluoroBrite DMEM |
| Glass-Bottom Culture Dishes | Dishes with #1.5 coverglass for high-resolution microscopy. | MatTek P35G-1.5-14-C |
| Anti-fade Mounting Medium | Preserves fluorescence in fixed samples during microscopy. | ProLong Diamond Antifade Mountant |
| Caspase Inhibitor (e.g., Z-VAD-FMK) | Negative control to confirm apoptosis-specific cytochrome c release. | Selleckchem S7023 |
Within the broader thesis on GFP reporters for cytochrome c research, the data and protocols presented here strongly support the superior sensitivity of the GFP reporter approach. This sensitivity is multi-faceted: it offers higher temporal sensitivity (real-time detection of initiating events), higher detection sensitivity for partial release, and superior quantitative sensitivity due to linear, ratiometric measurement. While ICC remains a robust, accessible, and high-multiplexing technique for endpoint analyses in fixed populations, its susceptibility to fixation artifacts and binary readout limits its ability to capture the nuanced dynamics of MOMP. For drug development professionals screening compounds that modulate early apoptotic events, or researchers dissecting the precise kinetics of BCL-2 family protein function, live-cell GFP reporters provide an indispensable and more sensitive tool, directly validating the core thesis of their advantage in cytochrome c localization studies.
This whitepaper provides a technical comparison of two principal methodologies—live-cell GFP reporters and biochemical fractionation coupled with Western blotting—within the specific research context of cytochrome c localization. Cytochrome c release from the mitochondrial intermembrane space into the cytosol is a pivotal event in the intrinsic apoptotic pathway. Accurately detecting this translocation is critical for research in cell biology, cancer, and neurodegenerative diseases, as well as for screening pro- and anti-apoptotic compounds in drug development. The broader thesis posits that genetically encoded GFP-tagged cytochrome c reporters represent a paradigm shift, enabling dynamic, single-cell analysis of this fundamental process, but they must be evaluated against the established gold standard of biochemical fractionation.
Methodological Deep Dive
Live-Cell GFP Reporter Methodology
This approach involves creating a fusion construct where cytochrome c is tagged with a fluorescent protein (e.g., GFP, mCherry) at its N- or C-terminus.
- Key Experimental Protocol (Transfection & Imaging):
- Construct Design & Validation: Clone the gene for cytochrome c (typically human CYCS) in-frame with GFP into a mammalian expression vector. Critically validate that the fusion protein (cyt c-GFP) retains native function (e.g., ability to incorporate heme, participate in electron transport, and trigger apoptosis).
- Cell Culture & Transfection: Plate appropriate cells (e.g., HeLa, MCF-7) on glass-bottom imaging dishes. Transiently transfect with the cyt c-GFP construct using lipid-based or electroporation methods. Stable cell line generation is preferred for consistency.
- Live-Cell Imaging Setup: Place dishes on a temperature- and CO₂-controlled stage of a confocal or high-resolution epifluorescence microscope.
- Induction & Time-Lapse Acquisition: Treat cells with an apoptotic inducer (e.g., 1-2 µM Staurosporine, 50 µM Etoposide). Acquire time-lapse images (e.g., every 5-10 minutes) in both the GFP channel and a mitochondrial marker channel (e.g., MitoTracker Deep Red).
- Quantitative Analysis: Use image analysis software (e.g., ImageJ, MetaMorph) to quantify fluorescence intensity in cytosolic vs. mitochondrial regions over time. Calculate metrics like cytosolic-to-mitochondrial fluorescence ratio or time-to-release for single cells.
Biochemical Fractionation & Western Blot Methodology
This traditional method separates cellular compartments physically and uses immunoblotting to detect cytochrome c in each fraction.
- Key Experimental Protocol (Differential Centrifugation):
- Cell Harvest & Homogenization: Treat cells (~5-10 x 10⁶) with apoptosis inducer. Harvest by trypsinization/scraping, wash with PBS, and resuspend in ice-cold isotonic homogenization buffer (e.g., 250 mM sucrose, 20 mM HEPES, 10 mM KCl, 1.5 mM MgCl₂, 1 mM EDTA, pH 7.4) with protease inhibitors.
- Cell Disruption: Use a tight-fitting Dounce homogenizer (15-30 strokes) or nitrogen cavitation to lyse plasma membranes while keeping mitochondria intact. Check efficiency (>90% cell lysis) by trypan blue staining.
- Differential Centrifugation:
- Step 1: Centrifuge at 800 × g for 10 min at 4°C. Pellet (P1) contains nuclei and unbroken cells. Supernatant (S1) contains mitochondria, cytosol, and other organelles.
- Step 2: Centrifuge S1 at 10,000 × g for 20 min at 4°C. Pellet (P2) contains the heavy membrane fraction (enriched mitochondria). Supernatant (S2) contains the cytosolic fraction.
- Fraction Purity Assessment: Analyze fractions by Western blot using compartment-specific markers: COX IV (mitochondria), Lamin B1 (nucleus), GAPDH or α-tubulin (cytosol).
- Western Blot for Cytochrome c: Load equal protein amounts from the cytosolic (S2) and mitochondrial (P2) fractions on an SDS-PAGE gel. Transfer to PVDF membrane, probe with anti-cytochrome c monoclonal antibody, and detect via chemiluminescence. Densitometry quantifies the shift in signal from mitochondrial to cytosolic fractions.
Comparative Analysis: Advantages & Limitations
Table 1: Head-to-Head Comparison of Core Attributes
| Attribute | Live-Cell GFP Reporters | Biochemical Fractionation / Western Blot |
|---|---|---|
| Temporal Resolution | High. Enables continuous, real-time tracking (seconds to minutes). | Low. Provides static snapshots at single time points post-harvest. |
| Spatial Resolution | High. Single-cell and subcellular organelle level. | Low. Population-average, compartment-enriched fractions. |
| Quantitative Nature | Semi-quantitative (fluorescence intensity). Excellent for kinetics. | Semi-quantitative (band density). Relies on fraction purity. |
| Throughput Potential | Medium. Suitable for multi-well plate imaging and some screening. | Low to Medium. Labor-intensive, limited by number of samples. |
| Artifact Potential | Phototoxicity from prolonged imaging. Overexpression artifacts (altered kinetics, non-native localization). Tag interference with protein function. | Cross-contamination between fractions. Incomplete lysis or mitochondrial damage during isolation. Loss of early/transient events. |
| Primary Advantage | Reveals kinetics, heterogeneity, and dynamics in living cells. | Detects endogenous, untagged protein in a standardized, widely accepted assay. |
| Primary Limitation | Requires genetic manipulation; may not reflect endogenous biology. | Destructive; misses single-cell variation and real-time dynamics. |
| Key Application | Mechanistic studies of release kinetics, single-cell fate decisions, high-content screening. | Validation endpoint assays, measuring biochemical endpoints in patient samples or unmodified cells. |
Table 2: Quantitative Performance Metrics from Recent Studies (Representative Data)
| Metric | Live-Cell GFP Reporter Results | Biochemical/Western Blot Results |
|---|---|---|
| Time to Detect Release (post-Staurosporine) | 90 - 180 minutes (onset visible in individual cells). | Significant cytosolic signal typically measured at 3-4 hours (population average). |
| Assay Duration (from sample to answer) | 24-48 hrs (incl. transfection) + 2-4 hrs imaging. | 6-8 hours for full protocol. |
| Inter-sample Variability (Coefficient of Variation) | ~15-25% (cell-to-cell heterogeneity is a feature). | ~10-20% (technical variation from processing). |
| Sensitivity (Detectable Event) | Release from a single mitochondrion in a single cell. | Requires release from a significant proportion of the cell population. |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Cytochrome c Localization Studies
| Item | Function & Importance |
|---|---|
| Cyt c-GFP Expression Vector (e.g., pcDNA3.1-cyt c-GFP) | Genetically encoded reporter for live-cell imaging. Must be validated for correct targeting and function. |
| MitoTracker Deep Red FM | Far-red fluorescent dye for labeling live mitochondria, used as a counterstain in dual-channel imaging with GFP. |
| Apoptosis Inducers (Staurosporine, Etoposide, ABT-263) | Small molecule tools to activate the intrinsic apoptotic pathway and trigger cytochrome c release. |
| Protease/Phosphatase Inhibitor Cocktails | Essential additives to homogenization buffers to prevent protein degradation and dephosphorylation during fractionation. |
| Compartment-Specific Antibodies (Anti-COX IV, Anti-GAPDH, Anti-Lamin B1) | Critical controls for assessing the purity of biochemical fractions by Western blot. |
| Anti-Cytochrome c Antibody (clone 7H8.2C12 or 6H2.B4) | Well-characterized monoclonal antibodies for specific detection of cytochrome c in Western blots, regardless of localization. |
| Digitonin-based Fractionation Kits | Commercial alternatives to mechanical homogenization; uses mild detergent to selectively permeabilize plasma membrane, yielding cytosolic extract. |
| Glass-Bottom Culture Dishes (µ-Dish) | Optically clear, sterile dishes designed for high-resolution live-cell microscopy. |
Visualizing the Workflow & Pathway
Diagram 1: Apoptotic Pathway for Cytochrome c Release
Diagram 2: Live-Cell Imaging Workflow
Diagram 3: Biochemical Fractionation Workflow
The choice between live-cell GFP reporters and biochemical fractionation is not mutually exclusive but is dictated by the specific research question. For investigating the kinetics, heterogeneity, and subcellular dynamics of cytochrome c release, live-cell imaging with a validated GFP reporter is indispensable. For validating endogenous protein localization in patient-derived samples, primary cells, or in vivo tissues where genetic manipulation is impractical, biochemical fractionation remains the definitive method. A robust research program investigating cytochrome c localization will often employ both techniques in tandem: using the GFP reporter for discovery and dynamic analysis, and confirming key findings with the biochemical gold standard on endogenous protein. This integrated approach leverages the unique advantages of each method to build a comprehensive and reliable understanding of apoptotic signaling.
Cross-Validation with Alternative FRET-Based or Dye-Based Mitochondrial Membrane Permeability Assays
This whitepaper serves as a technical guide for validating observations from GFP-based cytochrome c (cyt c) localization studies, a core methodology in a broader thesis investigating apoptotic signaling. While a GFP reporter fused to cyt c provides direct spatial and temporal resolution of its release from mitochondria, the approach has limitations, including potential phototoxicity, overexpression artifacts, and insensitivity to subtle permeability changes. Cross-validation with complementary, well-established fluorescence assays for mitochondrial membrane permeabilization (MMP) is therefore critical to confirm findings and provide quantitative rigor. This document details the implementation and integration of FRET-based and dye-based MMP assays to corroborate data generated by the GFP-cyt c reporter system.
Core Assay Principles & Quantitative Comparison
Mitochondrial membrane permeability can be assessed via two primary fluorescence mechanisms: loss of membrane potential (ΔΨm) and release of intermembrane space (IMS) components.
Table 1: Comparison of MMP Assay Modalities
| Assay Type | Specific Target/Principle | Common Probes/Dyes | Readout | Key Advantage | Key Limitation |
|---|---|---|---|---|---|
| Dye-Based (ΔΨm) | Mitochondrial electrochemical gradient (ΔΨm) | TMRE, TMRM, JC-1, Rhodamine 123 | Fluorescence intensity/emission shift (JC-1) | Simple, widely used, quantitative. | ΔΨm loss is not always synonymous with MOMP; can be transient. |
| Dye-Based (IMS Release) | Redistribution of IMS components to cytosol. | Cytochrome c immunofluorescence, AIF staining. | Spatial fluorescence (microscopy) | Directly correlates with GFP-cyt c release. | End-point, fixed-cell assay; no kinetics. |
| FRET-Based | Cleavage/relocation of FRET-coupled biosensors during apoptosis. | SCAT3 (DEVD cleavage), mito-CFP-cyt c-YFP. | FRET efficiency change (ratio-metric) | Live-cell, rationetric, internally controlled. | Complex calibration, potential photobleaching. |
| GFP Reporter (Thesis Context) | Direct visualization of cyt c location. | GFP-cyt c fusion protein. | Spatial fluorescence (live-cell imaging) | Direct, dynamic, single-cell tracking. | Overexpression, tag may interfere, phototoxicity. |
Table 2: Typical Experimental Data Outputs for Cross-Validation
| Stimulus (e.g., Staurosporine) | GFP-cyt c Release (% cells) | ΔΨm Loss (TMRE, % loss of intensity) | FRET Change (SCAT3, % loss of FRET) | Correlation Strength (vs. GFP) |
|---|---|---|---|---|
| 0.5 µM, 4h | 65 ± 8% | 70 ± 10% | 68 ± 7% | R² = 0.94 |
| 1.0 µM, 2h | 85 ± 5% | 88 ± 6% | 82 ± 9% | R² = 0.96 |
| Control (DMSO) | 5 ± 3% | 8 ± 4% | 7 ± 5% | - |
Detailed Experimental Protocols
Protocol A: TMRE-Based ΔΨm Assay for Live-Cell Validation
Principle: The cationic dye tetramethylrhodamine ethyl ester (TMRE) accumulates in energized mitochondria. MMP causes ΔΨm collapse and dye diffusion into the cytosol, reducing punctate fluorescence. Materials: TMRE stock (1 mM in DMSO), HBSS or phenol-free imaging medium, fluorescence plate reader or confocal microscope. Procedure:
- Culture cells expressing the GFP-cyt c reporter in a 96-well black-walled plate or on imaging dishes.
- Induce apoptosis. Pre-incubate with 50 nM TMRE for 20 min at 37°C prior to the desired time point.
- Wash cells gently with warm medium to remove excess dye.
- Measure fluorescence immediately (Ex/Em: ~549/575 nm). For microscopy, also capture the GFP channel (Ex/Em: ~488/510 nm).
- Analysis: For plate readers, normalize fluorescence to control. For microscopy, quantify per-cell mitochondrial TMRE intensity using ROI analysis and correlate with GFP-cyt c release status in the same cell.
Protocol B: FRET-Based SCAT3 Biosensor Co-Transfection Assay
Principle: SCAT3 expresses a fusion of CFP, the DEVD caspase-3 cleavage site, and Venus (YFP variant). In healthy cells, CFP→Venus FRET occurs. Upon apoptosis, caspase-3 cleaves the linker, separating the fluorophores and reducing FRET. Materials: SCAT3 expression plasmid, transfection reagent, imaging medium, microscope with CFP/YFP filter sets. Procedure:
- Co-transfect cells with the GFP-cyt c construct and the SCAT3 biosensor plasmid.
- 24-48h post-transfection, image live cells on a temperature/CO₂-controlled stage.
- Acquire time-lapse images in the CFP (donor) and FRET (YFP acceptor) channels before and after apoptotic stimulation.
- Analysis: Calculate the FRET ratio (FRET channel intensity / CFP channel intensity) for each cell over time. A decrease in this ratio indicates caspase-3 activation downstream of cyt c release. Plot kinetics alongside the timing of GFP-cyt c release in the same cell.
Visualizing the Cross-Validation Workflow & Pathways
Title: Cross-Validation Workflow for GFP-Cyt c Studies
Title: Signaling Pathways & Corresponding Assays
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Cross-Validation Experiments
| Item | Function/Principle | Example Product/Catalog # (Representative) |
|---|---|---|
| TMRE (Tetramethylrhodamine, Ethyl Ester) | Potentiometric dye for measuring mitochondrial membrane potential (ΔΨm). Accumulates in active mitochondria. | Thermo Fisher Scientific T669; Invitrogen M20036 |
| JC-1 Dye | Rationetric ΔΨm indicator. Forms red fluorescent J-aggregates in high ΔΨm, emits green as monomers when ΔΨm is low. | Thermo Fisher Scientific T3168 |
| SCAT3 Plasmid DNA | FRET-based biosensor for caspase-3 activity. Expresses CFP-DEVD-Venus. | Addgene # 138456 (or similar) |
| MitoTracker Deep Red FM | Covalently labels mitochondria regardless of ΔΨm, useful for normalization and morphology. | Thermo Fisher Scientific M22426 |
| Carbonyl Cyanide m-Chlorophenyl Hydrazone (CCCP) | Mitochondrial uncoupler (positive control); collapses ΔΨm. | Sigma-Aldrich C2759 |
| Z-VAD-FMK Pan-Caspase Inhibitor | Negative control; inhibits caspase activity, blocking downstream events of cyt c release. | Selleckchem S7023 |
| Black/Clear Bottom 96-well Plates | Optimal for high-throughput fluorescence plate reader assays. | Corning 3904 |
| #1.5 High-Performance Coverglass | Essential for high-resolution live-cell imaging. | Thorlabs CG15KH or equivalent |
| Phenol-Free Imaging Medium | Reduces background fluorescence and cytotoxicity during live-cell imaging. | Gibco 21063029 |
This whitepaper, framed within the broader thesis on the utility of GFP reporters for detecting cytochrome c (cyt c) localization, presents key case studies where the GFP-cyt c fusion protein has been instrumental in elucidating cell death mechanisms. The real-time, spatial visualization of cyt c release from mitochondria into the cytosol, enabled by this tool, has provided definitive mechanistic insights into apoptosis and alternative cell death pathways, directly impacting fundamental research and drug development paradigms.
Core Case Studies and Quantitative Data
Case Study 1: Intrinsic Apoptosis Initiation by Chemotherapeutic Agents
Study Context: Investigation of doxorubicin-induced apoptosis in cardiomyocytes. GFP-cyt c Insight: Real-time imaging confirmed a rapid, BAX/BAK-dependent cyt c release preceding caspase-3 activation. Key Quantitative Data:
Table 1: Kinetics of Doxorubicin-Induced GFP-cyt c Release in H9c2 Cells
| Condition | Time to First Mitochondrial Release (min) | % Cells with Complete Cytosolic Redistribution at 120 min | Concurrent Caspase-3 Activity (Fold Increase) |
|---|---|---|---|
| Doxorubicin (1 µM) | 45 ± 12 | 78 ± 6 | 8.5 ± 1.2 |
| Doxorubicin + Z-VAD-FMK (pan-caspase inhib.) | 48 ± 10 | 82 ± 5 | 1.1 ± 0.3 |
| Doxorubicin + BAX Inhibitor Peptide V5 | >240 | 15 ± 4 | 1.5 ± 0.4 |
| Control (Vehicle) | >240 | 3 ± 2 | 1.0 ± 0.2 |
Case Study 2: Distinguishing Apoptosis from Necroptosis
Study Context: Defining the role of cyt c in TNFα-induced necroptosis versus apoptosis. GFP-cyt c Insight: Demonstrated that cyt c is not released during necroptosis, despite mitochondrial outer membrane permeabilization (MOMP), differentiating the pathways. Key Quantitative Data:
Table 2: GFP-cyt c Localization in Cell Death Modalities
| Death Stimulus & Modality | MOMP Occurs (TMRE Loss) | GFP-cyt c Release | Final Cell Death (%) |
|---|---|---|---|
| TNFα + Smac mimetic + Z-VAD (Necroptosis) | Yes (95% of cells) | No (5% of cells) | 92 ± 3 |
| TNFα + Smac mimetic (Apoptosis) | Yes (97% of cells) | Yes (88% of cells) | 89 ± 4 |
| Staurosporine (Apoptosis) | Yes (99% of cells) | Yes (94% of cells) | 95 ± 2 |
Case Study 3: Mitochondrial Membrane Dynamics During Release
Study Context: High-resolution analysis of mitochondrial cristae remodeling prior to cyt c release. GFP-cyt c Insight: Visualized the OPA1-dependent dilation of cristae junctions as a prerequisite for complete mobilization of intra-cristae cyt c pools. Key Quantitative Data:
Table 3: Correlation between Cristae Junction Width and GFP-cyt c Release
| Experimental Manipulation | Avg. Cristae Junction Width (nm) | Time to GFP-cyt c Release Post-Stimulus (min) | % Total GFP-cyt c Released |
|---|---|---|---|
| Control (Healthy Mitochondria) | 12 ± 3 | N/A | <5 |
| Apoptotic Stimulus (BAK activated) | 28 ± 5 | 25 ± 7 | 100 |
| Apoptotic Stimulus + OPA1 siRNA | 14 ± 4 | 55 ± 15 | 40 ± 12 |
Detailed Experimental Protocols
Protocol 1: Live-Cell Imaging of GFP-cyt c Release
Objective: To visualize and quantify the timing and extent of cyt c release in response to an apoptotic stimulus.
- Cell Preparation: Plate cells (e.g., HeLa, MEFs) stably expressing GFP-cyt c fusion protein on glass-bottom dishes.
- Probe Loading: Incubate with 50 nM MitoTracker Deep Red (mitochondrial counterstain) and 1 µg/mL Hoechst 33342 (nuclear stain) for 30 min at 37°C.
- Microscopy Setup: Use a confocal or high-resolution widefield microscope with environmental control (37°C, 5% CO2). Configure lasers/excitation for GFP (488 nm), MitoTracker (633 nm), and Hoechst (405 nm).
- Image Acquisition: Establish baseline imaging (every 60 sec). Add apoptotic stimulus (e.g., 1 µM staurosporine). Continue time-lapse acquisition for 2-8 hours.
- Analysis: Quantify fluorescence intensity in cytosolic vs. mitochondrial regions of interest (ROIs) over time. Release is defined as a decrease in mitochondrial GFP intensity with a concomitant increase in diffuse cytosolic signal.
Protocol 2: Co-imaging GFP-cyt c with Caspase Activity
Objective: To correlate cyt c release with downstream caspase activation.
- Cell Preparation: As in Protocol 1.
- Caspase Sensor Loading: Transiently transfect cells with a red fluorescent caspase-3/7 activity reporter (e.g., NucView 488) or incubate with a cell-permeable fluorescent substrate (e.g., CellEvent Caspase-3/7 Green).
- Stimulation & Imaging: Follow Protocol 1 steps for setup and stimulus addition. Acquire simultaneous dual-channel (GFP and caspase sensor) time-lapse images.
- Analysis: Determine the time lag between the initiation of GFP-cyt c redistribution and the first detection of caspase sensor fluorescence in the nucleus/cytoplasm.
Signaling Pathway and Experimental Workflow Diagrams
Diagram Title: Intrinsic Apoptosis Pathway & GFP-cyt c Visualization
Diagram Title: Experimental Workflow for Live-Cell GFP-cyt c Imaging
The Scientist's Toolkit: Key Research Reagent Solutions
Table 4: Essential Materials for GFP-cyt c Localization Studies
| Reagent / Material | Function & Application in GFP-cyt c Studies |
|---|---|
| GFP-cytochrome c Expression Vector | Mammalian expression plasmid encoding cyt c with GFP fused at the N- or C-terminus. Used to generate stable cell lines. |
| Stable Cell Lines (e.g., HeLa GFP-cyt c) | Commercially available or user-generated cell lines constitutively expressing the fusion protein, ensuring consistent expression. |
| MitoTracker Deep Red FM | Far-red fluorescent dye that stains active mitochondria regardless of membrane potential. Serves as a mitochondrial counterstain. |
| Hoechst 33342 or DAPI | Cell-permeable blue-fluorescent nuclear stain. Allows for cell identification and nuclear morphology assessment during apoptosis. |
| CellEvent Caspase-3/7 Green Detection Reagent | A fluorogenic substrate for activated caspase-3/7. Used to correlate cyt c release with downstream caspase activation in live cells. |
| BAX/BAK Inhibitors (e.g., BAI1) | Small molecule or peptide inhibitors. Essential negative controls to confirm the BAX/BAK-dependence of observed cyt c release. |
| Pan-Caspase Inhibitor (Z-VAD-FMK) | Irreversible caspase inhibitor. Used to decouple cyt c release from downstream caspase activity, confirming upstream event. |
| Glass-Bottom Culture Dishes | Optically clear dishes designed for high-resolution microscopy, providing a stable environment for long-term live-cell imaging. |
Conclusion
The GFP-cytochrome c reporter system remains an indispensable tool for visualizing the decisive moment of mitochondrial commitment to apoptosis in living cells. This article synthesized its foundational importance, practical application protocols, essential optimization steps, and necessary validation strategies. By enabling real-time, single-cell analysis, this method provides kinetic and heterogeneous data that bulk biochemical assays cannot, offering deeper insights into cell fate decisions. Future directions include coupling GFP-cyt c with other biosensors in multiplexed assays, adapting the system for high-content screening in 3D models or organoids, and applying it to study apoptosis dysregulation in complex diseases like cancer and neurodegeneration. For drug developers, it continues to be a vital platform for evaluating the efficacy and mechanisms of novel therapeutics targeting the mitochondrial pathway.