Transmission Electron Microscopy for Early Apoptosis Identification: A Comprehensive Guide for Biomedical Research
This article provides a detailed guide for researchers and drug development professionals on utilizing Transmission Electron Microscopy (TEM) for the precise identification of early apoptotic cells.
Transmission Electron Microscopy for Early Apoptosis Identification: A Comprehensive Guide for Biomedical Research
Abstract
This article provides a detailed guide for researchers and drug development professionals on utilizing Transmission Electron Microscopy (TEM) for the precise identification of early apoptotic cells. It covers the foundational ultrastructural hallmarks of apoptosis, step-by-step methodological protocols for sample preparation and imaging, solutions for common troubleshooting scenarios, and a comparative analysis with other biochemical techniques. By synthesizing current standards and emerging applications, this resource aims to enhance the accuracy and reliability of apoptosis detection in experimental and clinical contexts, ultimately supporting advancements in disease research and therapeutic development.
The Gold Standard: Foundational Principles of Apoptosis and TEM Ultrastructure
Apoptosis, or programmed cell death, is a genetically determined process crucial for normal cell turnover, proper immune system function, embryonic development, and chemical-induced cell death [1]. Since its first formal description by Kerr, Wyllie, and Currie in 1972, apoptosis has been recognized as a distinct form of cell death characterized by specific morphological features and energy-dependent biochemical mechanisms [1] [2]. This in-depth technical guide focuses on defining early apoptosis, with particular emphasis on its identification using transmission electron microscopy (TEM), a method that provides unparalleled detail of the subcellular changes that occur during the initial phases of this process. For researchers investigating cell death, particularly in the context of drug development, accurate morphological assessment remains a cornerstone for defining apoptotic events, as biochemical analyses alone can sometimes yield false negatives due to cell-type-specific differences in DNA fragmentation [3].
Core Concepts and Definitions
Apoptosis vs. Necrosis: A Morphological Continuum
A fundamental understanding of apoptosis requires distinguishing it from necrosis. Apoptosis is an active, highly regulated process of programmed cell death, whereas necrosis is considered a passive, toxic process resulting from acute cellular injury, often following an energy-independent mode of death [1] [2]. The morphological and physiological differences between these two processes are critical for accurate identification.
However, it is increasingly evident that apoptosis and necrosis represent two extremes of a cell death continuum, and features of both may coexist in the same cell, especially in response to varying doses of the same stimulus [1] [3]. Factors such as the extent of ATP depletion and caspase availability can convert an ongoing apoptotic process into a necrotic one [1].
Table 1: Key Morphological Differences Between Apoptosis and Necrosis
| Feature | Apoptosis | Necrosis |
|---|---|---|
| Cellular Scope | Single cells or small clusters of cells [1] | Often contiguous cells, large fields [1] |
| Cell Size and Shape | Cell shrinkage and convolution [1] [3] | Cell swelling [1] |
| Nucleus | Pyknosis (condensation) and karyorrhexis (fragmentation) [1] [2] | Karyolysis, pyknosis, and karyorrhexis [1] |
| Cell Membrane | Intact until late stages, blebbing [1] [2] | Disrupted integrity early on [1] |
| Cytoplasmic Fate | Retained in apoptotic bodies [1] [2] | Released into the extracellular space [1] |
| Inflammatory Response | Essentially none [1] | Usually present [1] |
| Clearance Mechanism | Phagocytosis by macrophages or adjacent cells [1] [2] | Inflammatory cell recruitment [1] |
The Role of Apoptosis in Health and Disease
Apoptosis is a vital component of numerous physiological processes, including normal cell turnover, hormone-dependent atrophy, and embryonic development—such as the separation of fingers and toes in a developing human embryo [1] [2]. Inappropriate apoptosis, either excessive or insufficient, is a factor in many human diseases. Excessive apoptosis is implicated in neurodegenerative diseases and ischemic damage, while defective apoptosis can lead to uncontrolled cell proliferation, such as cancer [1] [2]. The ability to modulate cell death is therefore recognized for its immense therapeutic potential in drug development [1] [4].
Morphological Hallmarks of Early Apoptosis
Morphological assessment is one of the most definitive ways to identify and define apoptosis [3]. The early stages of apoptosis are characterized by a sequence of distinct structural changes that can be visualized with increasing detail through light, fluorescence, and electron microscopy.
Early-Stage Morphological Changes
The initial morphological signs of apoptosis become visible by both light and electron microscopy. The most characteristic early feature is chromatin condensation, known as pyknosis, where nuclear material aggregates peripherally under the nuclear membrane [1]. This is accompanied by overall cell shrinkage, where the cell becomes smaller in size, the cytoplasm becomes denser, and organelles are more tightly packed [1] [3]. Concurrently, the cell loses contact with its neighbours and the extracellular matrix, adopting a more rounded morphology [3]. This is followed by extensive plasma membrane blebbing, which occurs due to the activation of myosin light-chains and a rearrangement of the actin cytoskeleton, leading to the separation of the plasma membrane from the cytoskeleton [1] [3].
Advanced-Stage Morphological Changes
As apoptosis progresses, the nucleus undergoes karyorrhexis, or fragmentation into discrete bodies of condensed chromatin [1] [2]. The cell then separates into apoptotic bodies through a process called "budding." These apoptotic bodies are sealed membrane vesicles containing cytoplasm with tightly packed organelles, with or without nuclear fragments [1] [2]. These bodies are rapidly phagocytosed by macrophages or neighbouring cells, a process facilitated by surface changes on the apoptotic cells that prevent the release of cellular contents and thus an inflammatory response [1] [3].
Table 2: Summary of Key Morphological Events in Apoptosis
| Stage of Apoptosis | Key Morphological Event | Description |
|---|---|---|
| Early | Cell Shrinkage [1] [3] | Reduction in cell volume, denser cytoplasm, tightly packed organelles. |
| Early | Chromatin Condensation (Pyknosis) [1] [2] | Aggregation of nuclear material, often peripherally under the nuclear membrane. |
| Early | Membrane Blebbing [1] [2] | Formation of bulges in the plasma membrane due to cytoskeletal rearrangement. |
| Advanced | Nuclear Fragmentation (Karyorrhexis) [1] [2] | Breakdown of the nucleus into multiple fragments. |
| Advanced | Apoptotic Body Formation [1] [2] | Cell fragmentation into sealed vesicles containing cytoplasmic components and/or nuclear fragments. |
| Final | Phagocytosis [1] [2] | Engulfment and degradation of apoptotic bodies by phagocytes. |
Transmission Electron Microscopy for Identifying Early Apoptosis
Transmission electron microscopy (TEM) is considered a gold standard for the morphological assessment of apoptosis, as it allows for the detailed analysis of internal cellular structures and the definitive identification of hallmark features that are difficult to resolve with light microscopy [3].
Key Ultrastructural Features Resolved by TEM
TEM facilitates the visualization of critical early apoptotic events:
- Nuclear Changes: TEM can clearly reveal the peripheral aggregation of condensed chromatin in crescent-shaped masses against the intact nuclear membrane, a classic sign of early apoptosis [1] [3]. It can also distinguish the strong, organized chromatin compaction of caspase-dependent apoptosis from the lumpy, incomplete condensation seen in other death pathways [3].
- Cytoplasmic and Organellar Integrity: Unlike necrosis, where organelles swell and disintegrate, the integrity of cytoplasmic organelles is largely maintained during the early stages of apoptosis [1]. Mitochondria may appear condensed but remain intact in the initial phase before outer membrane permeabilization [2].
- Membrane Blebbing and Apoptotic Body Formation: The detailed structure of membrane blebs and the formation of sealed, intact apoptotic bodies are exquisitely resolved by TEM, confirming the regulated nature of the process [1] [3].
Experimental Protocol for TEM Assessment of Apoptosis
A detailed methodology for the morphological assessment of apoptosis via TEM is as follows [3]:
- Cell Fixation: Fix cells or tissue samples promptly. Primary fixation is typically performed using 2.5% glutaraldehyde in a 0.1 M sodium cacodylate buffer (pH 7.4) for a minimum of 1 hour at room temperature. This cross-links proteins and preserves ultrastructure.
- Post-Fixation: After washing with buffer, post-fix the samples with 1% osmium tetroxide in the same buffer for 1 hour. Osmium tetroxide stabilizes lipids and provides electron density to membranes.
- Dehydration: Dehydrate the fixed samples through a graded series of ethanol (e.g., 50%, 70%, 90%, 100%) to prepare for resin infiltration.
- Resin Infiltration and Embedding: Infiltrate the dehydrated samples with a resin, such as Spurr's or Epon-Araldite, and then embed them in fresh resin followed by polymerization at 60°C for 24-48 hours.
- Ultra-thin Sectioning: Use an ultramicrotome to cut the polymerized blocks into ultra-thin sections (typically 60-90 nm thick). Collect sections on copper or nickel grids.
- Staining: Stain the grids with heavy metals to enhance contrast. This typically involves uranyl acetate (for nucleic acids and proteins) followed by lead citrate (for general contrast).
- TEM Imaging and Analysis: Examine the stained sections under a transmission electron microscope operating at 60-80 kV. Systematically capture images at various magnifications to document and quantify apoptotic features. An initial screen at lower magnification can help identify areas of interest for more detailed analysis at higher magnifications.
Biochemical Pathways and Signaling Mechanisms
The morphological changes observed in apoptosis are the result of the activation of highly conserved biochemical pathways. The two best-understood activation mechanisms are the intrinsic and extrinsic pathways [2].
The Intrinsic (Mitochondrial) Pathway
The intrinsic pathway is activated by intracellular stress signals, such as DNA damage, radiation, oxidative stress, or growth factor withdrawal [1] [2]. These stresses lead to mitochondrial outer membrane permeabilization (MOMP), controlled by proteins of the Bcl-2 family, including the pro-apoptotic proteins Bax and Bak [2]. MOMP results in the release of cytochrome c and other proteins from the mitochondrial intermembrane space into the cytosol [2]. Cytochrome c binds to Apaf-1 and ATP to form the "apoptosome," a protein complex that activates the initiator caspase, pro-caspase-9 [2]. Activated caspase-9 then cleaves and activates the executioner caspase-3, which carries out the systematic degradation of cellular components, leading to the characteristic morphological changes of apoptosis [2].
The Extrinsic (Death Receptor) Pathway
The extrinsic pathway is initiated by the binding of extracellular death ligands (e.g., FasL, TNF-α) to cell-surface death receptors (e.g., Fas, TNFR1) [2]. This ligand-receptor binding leads to the formation of the Death-Inducing Signaling Complex (DISC), which recruits and activates the initiator caspase, pro-caspase-8 [2]. Activated caspase-8 can then directly cleave and activate executioner caspases like caspase-3, propagating the death signal [2].
Diagram 1: Core Apoptosis Signaling Pathways
The Scientist's Toolkit: Research Reagent Solutions
Accurate research into apoptosis, particularly its morphological assessment, relies on a suite of specialized reagents and tools.
Table 3: Essential Research Reagents for Apoptosis Morphology Studies
| Reagent / Tool | Primary Function in Apoptosis Research |
|---|---|
| Glutaraldehyde & Osmium Tetroxide [3] | Primary and post-fixatives for TEM; cross-link proteins and preserve ultrastructure, with osmium providing membrane contrast. |
| Uranyl Acetate & Lead Citrate [3] | Heavy metal stains for TEM grids; bind to cellular components (e.g., nucleic acids) to enhance electron scattering and image contrast. |
| Hematoxylin and Eosin (H&E) [3] | General purpose stains for light microscopy; hematoxylin stains nuclei blue, eosin stains cytoplasm pink, revealing pyknotic nuclei. |
| Hoechst 33342 / DAPI [3] | Fluorescent DNA-binding dyes for fluorescence microscopy; used to identify condensed and fragmented chromatin in apoptotic nuclei. |
| FRET-based Caspase Sensors [4] | Genetically encoded probes (e.g., ECFP-DEVD-EYFP) that lose FRET upon caspase cleavage; allow real-time, live-cell imaging of caspase activity. |
| Annexin V / Propidium Iodide (PI) [4] | Annexin V binds to phosphatidylserine externalized on early apoptotic cells; PI stains DNA in necrotic cells with permeable membranes. |
Advanced and Integrated Workflows
Modern research often integrates multiple techniques to confirm and quantify apoptosis. A powerful approach combines live-cell fluorescence imaging with subsequent TEM analysis. For instance, cells can be engineered to stably express a FRET-based caspase sensor and a mitochondrially-targeted fluorescent protein (e.g., Mito-DsRed) [4]. This allows for real-time discrimination in live cells: apoptotic cells show a loss of FRET (indicating caspase activation) while retaining mitochondrial fluorescence; necrotic cells lose the soluble FRET probe due to membrane rupture without a prior FRET change, but retain the mitochondrial marker [4]. Cells identified through this live-cell imaging can then be tracked and prepared for TEM to correlate the biochemical activity of caspases with the definitive ultrastructural morphology.
Diagram 2: Integrated Workflow for Apoptosis/Necrosis Discrimination
The precise definition of early apoptosis remains rooted in its distinct morphological hallmarks, with transmission electron microscopy providing the most definitive ultrastructural evidence. The characteristic sequence of cell shrinkage, chromatin condensation, membrane blebbing, and apoptotic body formation, as visualized by TEM, sets apoptosis apart from other forms of cell death like necrosis. For researchers and drug development professionals, a rigorous, morphology-centric approach, potentially enhanced by correlative live-cell and electron microscopy techniques, is essential for accurately identifying cell death mechanisms and evaluating the efficacy of therapeutic agents designed to modulate these critical pathways.
Transmission Electron Microscopy (TEM) stands as the undisputed gold standard for subcellular analysis, providing researchers with an unparalleled tool to visualize the intricate architecture of life at the nanoscale. While various microscopy techniques offer insights into cellular structures, TEM delivers exceptional resolution that enables the detailed examination of intracellular components and their alterations in response to pathological conditions. In the specific context of apoptosis research, TEM's capability to reveal definitive ultrastructural changes during programmed cell death makes it an indispensable technology for both basic research and drug development. The technique's extraordinary resolving power, capable of visualizing features as small as 0.1 nanometers, allows scientists to distinguish not only individual organelles but also subtle morphological transformations that signify early apoptotic events—information that remains inaccessible through other imaging modalities [5] [6].
The significance of TEM in apoptosis research extends beyond mere structural observation. As drug development increasingly focuses on targeted therapies that modulate programmed cell death pathways, the ability to precisely identify and validate apoptotic events becomes crucial. TEM provides this validation through direct visualization of characteristic morphological hallmarks, serving as a critical confirmatory tool alongside biochemical and molecular assays. For researchers investigating novel therapeutic agents designed to induce or inhibit apoptosis in cancer cells, neurodegenerative diseases, or other conditions, TEM offers the definitive evidence needed to understand mechanism of action at the cellular level [7] [8]. This article explores the technical foundations of TEM's superior capabilities, its specific applications in apoptosis detection, and the methodologies that make it an irreplaceable asset in modern cell biology research.
Technical Superiority: Resolution and Magnification
The exceptional capabilities of Transmission Electron Microscopy stem from fundamental physical principles that differentiate it from other microscopy techniques. Unlike light microscopes that use photons, TEM employs a beam of electrons accelerated under high voltage, typically ranging from 60-300 kV. The shorter effective wavelength of these electrons according to de Broglie's equation provides TEM with its extraordinary resolving power, enabling visualization of structures at the nanometer scale that are orders of magnitude smaller than what light-based microscopes can detect [6].
Table 1: Comparative Analysis of Microscopy Techniques
| Parameter | Transmission Electron Microscope (TEM) | Scanning Electron Microscope (SEM) | Light Microscope (LM) |
|---|---|---|---|
| Maximum Resolution | 0.1 nm | 10 nm | 200 nm |
| Maximum Magnification | >50,000,000x | 1-2,000,000x | ~1,500x |
| Optimal Specimen Thickness | <100 nm | No thickness restriction | 1-10 μm (sections) |
| Image Type | 2D projection of internal structure | 3D surface topography | 2D color image |
| Primary Applications in Biology | Intracellular ultrastructure, organelle morphology, viral identification | Surface features, cellular topography, cilia/flagella | Live cell imaging, histology, basic cytology |
This dramatic difference in resolution directly impacts the utility of each technique for apoptosis research. While light microscopy might reveal overall cellular shrinkage and light microscopes equipped with fluorescence capabilities can show phosphatidylserine externalization using Annexin V staining, only TEM can visualize the definitive ultrastructural changes that confirm apoptotic progression, including chromatin condensation into precisely defined geometric patterns, mitochondrial remodeling with cristae disruption, and the formation of apoptotic bodies with intact organelles [5] [6].
The fundamental distinction between resolution and magnification further underscores TEM's superiority. Magnification simply enlarges an image, while resolution determines the level of detail that can be discerned. TEM achieves both extreme magnification (exceeding 50 million times) and exceptional resolution simultaneously, enabling researchers to not just "zoom in" on subcellular structures but to actually resolve their fine details with clarity. This capability is particularly crucial for distinguishing early apoptotic changes from other forms of cell death, such as necrosis, which presents dramatically different ultrastructural features including plasma membrane rupture and organelle swelling without the organized chromatin condensation characteristic of apoptosis [9].
TEM in Apoptosis Research: Visualizing Cell Death Mechanisms
The application of TEM in apoptosis research provides unparalleled insights into the morphological manifestations of programmed cell death. Apoptosis occurs through two principal pathways—the extrinsic (death receptor) pathway and the intrinsic (mitochondrial) pathway—both culminating in characteristic structural changes that TEM visualizes with exceptional clarity [8]. In the intrinsic pathway, cellular stress factors cause mitochondrial outer membrane permeabilization, leading to cytochrome c release and activation of executioner caspases. The extrinsic pathway initiates through external death signals binding to surface receptors like Fas and TNF receptors, similarly activating caspase cascades. While biochemical assays can detect caspase activation and molecular techniques can identify gene expression changes, only TEM directly reveals the structural consequences of these molecular events [7] [8].
TEM enables researchers to identify the defining ultrastructural features of apoptosis, including:
- Chromatin condensation: Margination of nuclear material into dense, geometric masses adjacent to the nuclear envelope
- Nuclear fragmentation: Progressive convolution of the nuclear membrane followed by karyorrhexis (nuclear fragmentation)
- Cytoplasmic condensation: Compaction of cytoplasmic contents with organelle preservation
- Membrane blebbing: Formation of protuberances on the cell surface that evolve into apoptotic bodies
- Apoptotic body formation: Separation of cell fragments containing intact organelles and nuclear material
These morphological markers appear in specific temporal sequences, beginning with chromatin condensation and culminating in apoptotic body formation and phagocytosis by neighboring cells. TEM not only captures each stage in this process but can also reveal pathway-specific features, such as mitochondrial alterations in the intrinsic pathway including cristae disruption and matrix condensation [8].
For drug development professionals, these ultrastructural observations provide critical validation of therapeutic mechanisms. When evaluating novel compounds designed to induce apoptosis in cancer cells, TEM analysis offers definitive proof of efficacy at the cellular level. Similarly, in toxicology studies, TEM can identify unwanted apoptotic induction in non-target tissues, providing essential safety data. The technology's precision allows researchers to distinguish between complete apoptosis, incomplete apoptosis (abortive attempts), and alternative cell death modalities like autophagy, necroptosis, and pyroptosis, each of which presents distinct ultrastructural profiles [10].
Table 2: Key Ultrastructural Features of Apoptosis Visualized by TEM
| Apoptotic Stage | TEM-Detectable Features | Biological Significance |
|---|---|---|
| Early Phase | Chromatin margination, nucleolar disintegration, cytoplasmic compaction | Initial commitment to apoptotic pathway; potentially reversible |
| Intermediate Phase | Nuclear membrane convolution, organelle clustering, mitochondrial condensation | Irreversible progression; caspase activation |
| Advanced Phase | Nuclear fragmentation (karyorrhexis), pronounced membrane blebbing | Execution phase; widespread proteolytic activity |
| Terminal Phase | Apoptotic body formation, phagocytosis by adjacent cells | Clean elimination without inflammation |
Experimental Protocols for Apoptosis Detection Using TEM
Sample Preparation Workflow
Proper sample preparation is critical for successful TEM analysis of apoptotic cells. The multi-step process must preserve ultrastructural integrity while providing sufficient contrast for electron imaging:
Chemical Fixation: Primary fixation using 2-4% glutaraldehyde in 0.1M phosphate buffer (pH 7.4) for several hours at 4°C stabilizes cellular structures by cross-linking proteins. This initial fixation must occur rapidly after experimental induction of apoptosis to capture the desired stage of cell death. Post-fixation with 1-2% osmium tetroxide for 1-2 hours further stabilizes membranes and provides electron density [11].
Dehydration and Embedding: Sequential ethanol or acetone dehydration (30-100%) prepares samples for infiltration with epoxy resin (such as Epon or Araldite). Proper infiltration ensures uniform embedding and facilitates thin sectioning. Polymerization occurs at 60°C for 24-48 hours, producing blocks with hardened resin containing the fixed cells [11].
Ultramicrotomy: Using ultramicrotomes equipped with diamond knives, sections are cut at 60-90nm thickness. The precise thickness is critical—too thick sections reduce resolution, while too thin sections provide insufficient contrast. Sections are collected on copper or nickel grids (typically 200-300 mesh) for subsequent staining [11].
Contrast Enhancement: Heavy metal staining, typically with uranyl acetate (5-10% for 10-30 minutes) followed by lead citrate (1-2% for 1-5 minutes), binds to cellular components and increases electron scattering. This step dramatically improves contrast, making membranes, chromatin, and organelles clearly distinguishable [11].
Diagram 1: Sample preparation workflow for TEM analysis of apoptosis.
Image Acquisition and Analysis
Modern TEM imaging for apoptosis research combines conventional bright-field imaging with advanced techniques:
Conventional Imaging: Standard bright-field TEM operated at 60-100kV provides high-resolution overviews of cellular ultrastructure. Images are typically captured using digital CCD cameras with resolutions of 4k×4k pixels or higher. Multiple micrographs at increasing magnifications (from low-magnification surveys to high-magnification details) document both overall cellular changes and specific organelle alterations [11].
Quantitative Morphometry: Advanced TEM analysis includes morphometric assessment of apoptotic features using specialized software. Parameters commonly quantified include:
- Nuclear-to-cytoplasmic ratio changes
- Mitochondrial area and cristae density
- Apoptotic body count and size distribution
- Chromatin condensation patterns
Recent advancements integrate deep learning with TEM analysis, dramatically improving efficiency and reproducibility. Automated segmentation pipelines can reduce analysis time by up to 90% compared to manual methods while maintaining high accuracy. These systems employ probabilistic interactive segmentation models that leverage uncertainty analysis to identify regions requiring researcher attention, creating an efficient human-in-the-loop workflow [11].
Advanced TEM Techniques in Cell Death Research
Cryo-Electron Microscopy
Cryo-fixation techniques represent a significant advancement for apoptosis research. Instead of chemical fixation, high-pressure freezing rapidly vitrifies cellular water without ice crystal formation, preserving structures in a near-native state. Cryo-TEM enables imaging of unstained, frozen-hydrated specimens, revealing ultrastructural details without potential chemical artifacts. For apoptosis studies, this approach can capture very early membrane and organelle changes that might be altered by conventional processing [12].
Immunogold Labeling
TEM's capabilities extend beyond morphology to molecular localization through immunoelectron microscopy. This technique uses antibodies conjugated to colloidal gold particles (typically 5-20nm) to precisely localize specific antigens at the ultrastructural level. In apoptosis research, immunogold labeling can identify:
- Subcellular distribution of Bcl-2 family proteins
- Caspase activation and cleavage sites
- Death receptor clustering
- Phosphatidylserine translocation
Combining immunogold with conventional TEM provides correlative data linking molecular events with structural changes, offering comprehensive insights into apoptotic mechanisms [12].
Electron Tomography
Electron tomography reconstructs three-dimensional ultrastructure from TEM tilt series, providing volumetric data about apoptotic cells. This technique reveals spatial relationships between organelles during apoptosis, such as mitochondrial-ER contact sites during calcium signaling, and the precise architecture of the apoptosome complex. The 3D perspective helps researchers understand structural dynamics that may be misinterpreted in conventional 2D projections [12].
Research Reagent Solutions for TEM Apoptosis Studies
Table 3: Essential Reagents for TEM-Based Apoptosis Research
| Reagent/Category | Specific Examples | Function in TEM Workflow |
|---|---|---|
| Primary Fixatives | Glutaraldehyde, Paraformaldehyde | Cross-links proteins; stabilizes ultrastructure |
| Secondary Fixatives | Osmium Tetroxide | Preserves lipids; provides membrane contrast |
| Dehydration Media | Ethanol, Acetone | Removes water prior to resin infiltration |
| Embedding Resins | Epon, Araldite, LR White | Provides structural support for sectioning |
| Section Stains | Uranyl Acetate, Lead Citrate | Enhances electron contrast of cellular components |
| Grid Substrates | Copper, Nickel, Gold Grids | Supports ultra-thin sections during imaging |
| Immunolabeling Reagents | Protein A-Gold, Antibody-Gold Conjugates | Localizes specific antigens at ultrastructural level |
| Specialized Reagents | TUNEM for DNA fragmentation | Correlates biochemical and ultrastructural apoptosis markers |
Current Research and Future Perspectives
TEM technology continues to evolve, with recent advancements further solidifying its role as the gold standard for apoptosis research. The integration of machine learning algorithms for automated image analysis represents a particularly promising development. Deep learning frameworks now enable high-throughput segmentation and classification of apoptotic features in TEM images, achieving accuracy comparable to human experts while reducing analysis time by approximately 90% [11]. These systems utilize probabilistic models that generate multiple segmentation hypotheses and identify regions of uncertainty where researcher input is most valuable, creating an efficient collaborative workflow between human expertise and computational power.
In drug development, TEM remains indispensable for validating the mechanisms of novel therapeutic agents. Nanoparticle-based delivery systems designed to induce apoptosis in cancer cells require thorough characterization of their cellular interactions and effects, which TEM provides at unprecedented resolution [8] [10]. Similarly, the growing interest in alternative programmed cell death pathways—including pyroptosis, ferroptosis, and cuproptosis—relies on TEM for definitive identification, as each pathway presents distinct ultrastructural signatures [10]. The ability to distinguish between these modalities is crucial for understanding therapeutic efficacy and potential side effects.
Future directions in TEM apoptosis research include:
- Correlative Light and Electron Microscopy (CLEM): Combining live-cell imaging with subsequent TEM analysis to link dynamic processes with ultructural details
- Automated high-content screening: Applying machine learning to analyze large TEM datasets from drug screening assays
- Integrated molecular mapping: Combining TEM with elemental analysis through energy-dispersive X-ray spectroscopy (EDX) or electron energy loss spectroscopy (EELS)
- Cryo-electron tomography: Visualizing apoptotic structures in three dimensions at near-atomic resolution
These technological advances ensure that TEM will maintain its essential role in apoptosis research and drug development, providing the definitive standard against which other methods are measured. As nanomedicine continues to develop novel approaches to modulate cell death pathways, TEM's unparalleled resolution will remain critical for validating therapeutic mechanisms and optimizing treatment strategies [10].
Diagram 2: Key morphological events in apoptosis visualized by TEM.
Transmission electron microscopy (TEM) remains the gold standard for the definitive identification of early apoptotic cells, providing unparalleled resolution of the ultrastructural hallmarks that distinguish this programmed cell death from other forms of cellular demise. This technical guide details the core morphological features—chromatin margination, cytoplasmic condensation, and organelle integrity—within the context of contemporary apoptosis research. We provide a synthesized overview of the biochemical pathways initiating these changes, comprehensive protocols for TEM-based ultrastructural analysis, and a curated toolkit of research reagents. Designed for researchers and drug development professionals, this whitepaper serves as an authoritative resource for the precise identification and study of early apoptosis using transmission electron microscopy.
Apoptosis, or programmed cell death, is a genetically controlled process crucial for normal cell turnover, embryonic development, and proper immune system functioning [13] [14]. Unlike necrosis, which is an uncontrolled, inflammatory form of cell death characterized by cell swelling and membrane rupture, apoptosis is a silent, immunologically inert process that occurs without damaging neighbouring cells [15] [14]. The unique value of transmission electron microscopy in apoptosis research lies in its ability to visualize specific subcellular morphological changes that are considered the definitive hallmark of this process, allowing for its distinction from other regulated cell death pathways like necroptosis and pyroptosis [13] [14].
Early ultrastructural changes in apoptosis are triggered by the activation of a family of cysteine-aspartic proteases known as caspases. These caspases are synthesized as inactive procaspases and are activated through a proteolytic cascade that cleaves key cellular substrates, leading to the characteristic morphological alterations [14]. While various biochemical assays and light microscopy techniques exist for detecting apoptosis, TEM is uniquely capable of confirming the diagnosis by revealing the intricate subcellular details, such as nuclear fragmentation, apoptotic bodies, blebbing, and cytoplasmic or nuclear condensation, at the nanoscale level [16] [13]. This makes TEM an indispensable tool for validating findings from other methods and for conducting in-depth mechanistic studies of cell death.
Core Ultrastructural Features of Early Apoptosis
The early phase of apoptosis is marked by a conserved sequence of ultrastructural events, predominantly affecting the nucleus and the cytoplasm. The following features are consistently observed across cell types and apoptotic stimuli.
Chromatin Margination and Nuclear Degradation
The most diagnostic feature of early apoptosis is the remodeling of nuclear chromatin. This process begins with chromatin condensation, where the granular nuclear material becomes densely packed and electron-dense under TEM. This is rapidly followed by chromatin margination, a process where the condensed chromatin aggregates into sharply defined, coarse masses that abut the inner nuclear membrane [13]. The central nucleoplasm often appears comparatively empty or less electron-dense, creating a characteristic "halo" effect.
Subsequent nuclear changes include:
- Pyknosis: A reduction in nuclear volume, leading to a small, dense nucleus.
- Nuclear Membrane Irregularity: The nuclear envelope may become convoluted, wavy, or blebbed [13].
- Karyorrhexis: The eventual fragmentation of the nucleus into discrete, membrane-bound apoptotic bodies containing nuclear material [13].
Cytoplasmic Condensation and Organelle Integrity
Concurrent with nuclear changes, the cytoplasm undergoes profound restructuring. The cell commits to a reduction in volume, leading to increased cytoplasmic density. Despite this condensation, the integrity of major organelles is largely maintained in the early stages, a key feature distinguishing apoptosis from necrotic cell death.
Key cytoplasmic features include:
- Cytoplasmic Condensation: The entire cell shrinks, resulting in a darker, more compact cytoplasm in TEM images [13] [14].
- Organelle Preservation: Mitochondria, endoplasmic reticulum, and Golgi apparatus typically remain structurally intact, though they may be more tightly packed. A reduction in mitochondrial number and expansion of the rough endoplasmic reticulum are frequently observed [13].
- Membrane Blebbing: The plasma membrane undergoes dynamic "blebbing," forming outward protrusions that eventually separate from the cell. This is a result of actomyosin contraction mediated by the caspase-mediated activation of ROCK1 kinase [17] [18].
- Formation of Pinopode-like Projections: Thin, finger-like membrane protrusions are commonly observed, contributing to cell fragmentation [13].
Table 1: Quantitative Analysis of Ultrastructural Features in Early Apoptosis
| Ultrastructural Feature | Typical Appearance in Early Apoptosis | Key Distinguishing Factor from Necrosis |
|---|---|---|
| Chromatin | Margination, condensation, pyknosis | Swelling, karyolysis (dissolution) |
| Nucleus | Fragmentation into apoptotic bodies | Swelling, followed by rupture |
| Cytoplasm | Condensation, increased density | Swelling (oncosis), loss of structure |
| Plasma Membrane | Blebbing, intact integrity | Early rupture, loss of integrity |
| Organelles | Generally intact, but densely packed | Swelling, dilation, gross disruption |
| Cellular Volume | Decreased (cell shrinkage) | Increased (cell swelling) |
| Inflammatory Response | None (immunologically silent) | Significant (pro-inflammatory) |
The "Footprint of Death" and Apoptotic Body Formation
A recent discovery, termed the "FOotprint Of Death" (FOOD), describes a mechanism where adherent apoptotic cells retract and leave behind actin-rich membrane tracks on the substrate [17]. These footprints subsequently vesicularise into large apoptotic bodies (F-ApoEVs), which are ~2 μm in diameter and expose phosphatidylserine (PS) on their surface. This process, regulated by ROCK1, provides an alternative pathway for generating apoptotic bodies that mark the site of cell death and facilitate communication with phagocytes [17]. This finding expands the understanding of how ultrastructural changes are not merely a prelude to disintegration but are part of an active signaling process.
Biochemical Pathways Underlying Ultrastructural Changes
The dramatic morphological changes observed via TEM are the direct result of a tightly regulated biochemical cascade, primarily driven by caspase activation.
Diagram 1: Apoptotic Signaling to Ultrastructure
The intrinsic (mitochondrial) pathway is triggered by internal stressors like DNA damage, leading to mitochondrial outer membrane permeabilization and the release of cytochrome c. Cytochrome c, in combination with APAF-1, forms the apoptosome, which activates caspase-9. The extrinsic pathway is initiated by the ligation of death receptors on the cell surface, which recruit adaptor proteins to activate caspase-8. Both pathways converge on the activation of executioner caspases, primarily caspase-3 and -7 [14]. These executioner caspases then cleave over 600 cellular substrates, including key structural proteins.
The cleavage of proteins such as ROCK1 leads to the uncontrolled contraction of the actomyosin cortex, which is directly responsible for the cytoplasmic condensation and membrane blebbing observed under TEM [17] [18]. Simultaneously, caspase-activated endonucleases are responsible for the DNA fragmentation and chromatin changes that characterize the apoptotic nucleus.
Experimental Protocols for TEM-Based Apoptosis Identification
Sample Preparation and Induction of Apoptosis
A critical step in TEM analysis is the faithful preservation of cellular ultrastructure through meticulous sample preparation. The following protocol, adapted from studies on human lens epithelial cells and other models, provides a robust framework [13].
Cell Culture and Apoptosis Induction:
- Cell Lines: Common models include HeLa cells (human cervical adenocarcinoma), A431 (squamous epithelial), MEFs (mouse embryonic fibroblasts), and primary cells like HUVECs [17] [15].
- Apoptotic Inducers:
- BH3-mimetics: A cocktail of ABT-737 and S63845 to target the intrinsic pathway [17].
- Chemotherapeutic Agents: Doxorubicin (5 μmol/L), which induces DNA damage and the p53 pathway [15].
- Kinase Inhibitors: Staurosporine (10 μM), a broad-spectrum kinase inhibitor that triggers intrinsic apoptosis [16].
- Other Stimuli: UV irradiation, etoposide, or viral infection [17].
TEM Sample Preparation Workflow:
- Fixation: Immediately after apoptosis induction, fix cells or tissue specimens in a neutral-buffered 3% glutaraldehyde solution for at least 90 minutes. This cross-links proteins and preserves structure.
- Post-fixation: Wash the sample and post-fix in 1-2% osmium tetroxide (OsO4) for 1-2 hours. Osmium tetroxide stabilizes lipids and provides electron density to membranes.
- Dehydration: Gradually dehydrate the specimen using a graded ethanol series (e.g., 50%, 70%, 90%, 100%) to remove all water.
- Embedding: Infiltrate and embed the dehydrated sample in a resin, such as Epon 812, which is then polymerized into a hard block at high temperature.
- Sectioning: Use an ultramicrotome to cut ultrathin sections (60-80 nm) from the resin block. These golden-colored sections are collected on TEM grids.
- Staining: Stain the sections with heavy metals: first with uranyl acetate (for contrast of nucleic acids and proteins) and then with lead citrate (for general contrast enhancement).
Diagram 2: TEM Sample Preparation Workflow
Image Acquisition and Quantitative Analysis
Once prepared, grids are imaged using a transmission electron microscope (e.g., JEOL JEM-1011) operating at 80 kV [13]. For a comprehensive analysis, it is recommended to systematically image multiple areas of the sample. For instance, one study examined at least six different areas of anterior lens capsules, with at least 25 cells analyzed per case to ensure statistically significant identification of apoptotic events [13].
Quantitative analysis can include:
- Apoptotic Index: The percentage of cells displaying definitive ultrastructural features of apoptosis within a total cell count.
- Morphometry: Measuring the size of apoptotic cells, nuclear fragments, and apoptotic bodies using image analysis software.
- Frequency of Specific Features: Quantifying the prevalence of features like membrane blebs, pinopode-like projections, or specific organelle alterations.
Table 2: Research Reagent Solutions for TEM Apoptosis Analysis
| Reagent / Material | Function / Application | Technical Notes |
|---|---|---|
| Glutaraldehyde (3%) | Primary fixative; cross-links proteins to preserve ultrastructure. | Use a neutral, phosphate-buffered solution. |
| Osmium Tetroxide (1-2%) | Post-fixative; stabilizes phospholipids and adds electron density. | Highly toxic; requires use in a fume hood. |
| Epon 812 Resin | Embedding medium; provides support for ultrathin sectioning. | Infiltration must be gradual for proper specimen penetration. |
| Uranyl Acetate | Heavy metal stain; enhances contrast of nucleic acids and proteins. | Light-sensitive; often prepared in ethanol or methanol. |
| Lead Citrate | Heavy metal stain; provides general contrast enhancement. | Must be protected from atmospheric CO2 to prevent precipitate formation. |
| Staurosporine | Induces intrinsic apoptosis; positive control for experiments. | A broad-spectrum protein kinase inhibitor. [16] |
| Doxorubicin | Chemotherapeutic agent; induces DNA damage-mediated apoptosis. | Used at ~5 μmol/L concentration. [15] |
| BH3-mimetic Cocktail | Targets Bcl-2 proteins to specifically trigger the intrinsic pathway. | Contains ABT-737 and S63845. [17] |
Discussion and Technical Considerations
While TEM is the gold standard for morphological confirmation of apoptosis, several technical considerations must be acknowledged. The process of apoptosis in vivo is remarkably rapid, estimated to last from two to 24 hours, meaning only a few cells undergoing apoptosis may be present at a single time point [13]. This, combined with the limited number of cells that TEM can feasibly study in a single session, creates a risk of underestimation or false-negative results if sampling is not sufficiently thorough [19] [13]. Therefore, it is critical to analyze multiple sections and areas from each sample.
For a comprehensive research strategy, TEM should be integrated with other complementary techniques. Flow cytometry, for instance, excels at providing high-throughput, quantitative data on cell viability and can distinguish early apoptotic (e.g., Annexin V-positive) from late apoptotic and necrotic populations, but it lacks the ability to visualize ultrastructural details [20]. Fluorescence microscopy allows for real-time monitoring of apoptosis using probes for caspase activation or phosphatidylserine exposure, but it is limited by resolution and potential phototoxicity [16] [20]. Thus, a multi-modal approach, where TEM is used to validate and provide deeper insight into findings from these other methods, represents the most powerful strategy for apoptosis research.
The ultrastructural features of early apoptosis—chromatin margination, cytoplasmic condensation, and preserved organelle integrity—are definitive markers that can be unequivocally identified using transmission electron microscopy. The detailed protocols and analytical frameworks presented in this whitepaper provide researchers with the tools to reliably detect and characterize this fundamental biological process. As research continues to unveil novel aspects of apoptotic cell death, such as the formation of the "Footprint of Death" [17], the resolving power of TEM will remain indispensable for validating biochemical findings and advancing our understanding of cell death in health, disease, and therapeutic intervention.
Within the context of early apoptosis research, transmission electron microscopy (TEM) remains an indispensable tool for the precise identification and differentiation of cell death pathways. While molecular techniques have advanced significantly, ultrastructural criteria continue to provide the definitive standard for classifying apoptotic versus necrotic cell death, particularly in complex pathological contexts such as atherosclerosis where biochemical data alone may lead to misinterpretation [21]. The morphological features observable at the nanoscale level offer researchers unparalleled insight into the fundamental processes of cellular demise, enabling accurate discrimination between the highly organized program of apoptosis and the disruptive cascade of necrosis.
This technical guide provides a comprehensive framework for utilizing TEM in cell death analysis, detailing the characteristic ultrastructural features that distinguish different forms of cell death, with particular emphasis on the early morphological indicators that are crucial for accurate experimental interpretation in drug development and basic research.
Core Ultrastructural Features: A Comparative Analysis
Defining Morphological Characteristics
The ultrastructural hallmarks of apoptosis and necrosis manifest through distinct alterations in cellular and organellar architecture. The following table summarizes the key differentiating features:
Table 1: Ultrastructural Features of Apoptosis versus Necrosis
| Cellular Component | Apoptosis | Necrosis |
|---|---|---|
| Overall Cell Morphology | Cell shrinkage and rounding; preservation of membrane integrity until late stages [22] | Cell swelling; severe dilation of organelles; eventual membrane rupture [21] |
| Plasma Membrane | Membrane blebbing (zeiosis) and formation of apoptotic bodies [23] [22] | Rapid membrane rupture with content leakage; loss of adhesion structures [15] |
| Nucleus | Chromatin condensation (pyknosis) and marginalion; nuclear fragmentation (karyorrhexis) [24] | Pale nucleus with minimal chromatin condensation; eventual karyolysis [21] |
| Mitochondria | May appear relatively normal or condensed; cytochrome c release without gross swelling [25] | Severe swelling and dilation; rupture of mitochondrial membranes [21] |
| Other Organelles | Generally preserved structure with compaction [26] | Gross dilation of endoplasmic reticulum and other organelles [21] |
| Cellular Contents | Retained within membrane-bound bodies | Released into extracellular space |
The progression of these two forms of cell death follows fundamentally different sequences, as illustrated below:
Temporal Dynamics and Detection Windows
The sequence of ultrastructural events follows distinct temporal patterns that are critical for accurate identification. In apoptosis, chromatin condensation typically represents one of the earliest detectable morphological changes, followed by cytoplasmic compaction and membrane blebbing [24]. These changes occur while membrane integrity remains largely intact. In contrast, necrosis initiates with organellar swelling, particularly affecting mitochondria and endoplasmic reticulum, progressing rapidly to plasma membrane rupture [21]. This fundamental difference in progression—controlled dismantling versus catastrophic failure—forms the basis for ultrastructural discrimination.
Experimental Protocols for Ultrastructural Analysis
Sample Preparation for Transmission Electron Microscopy
Proper sample preparation is paramount for preserving the delicate ultrastructural features that distinguish apoptosis from necrosis. The following protocol outlines the standardized methodology for TEM-based cell death analysis:
Primary Fixation: Immerse cell pellets or tissue samples immediately in 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer (pH 7.4) for a minimum of 2 hours at 4°C. For tissue samples, perfuse fixation is recommended when possible [21]. This rapid fixation is critical for preserving the native cellular architecture and preventing post-mortem artifacts.
Secondary Fixation: Post-fix samples in 1% osmium tetroxide in the same buffer for 1-2 hours at room temperature. This secondary fixation stabilizes lipid membranes and provides electron scattering contrast.
Dehydration: Subject samples to a graded ethanol series (50%, 70%, 90%, 100%) followed by propylene oxide to ensure complete dehydration while minimizing structural collapse.
Embedding and Polymerization: Infiltrate samples with epoxy resin (such as Epon or Araldite) and polymerize at 60°C for 48 hours. This process provides structural support for ultra-thin sectioning.
Sectioning and Staining: Cut ultrathin sections (60-90nm) using an ultramicrotome equipped with a diamond knife. Mount sections on copper grids and stain with uranyl acetate and lead citrate to enhance contrast for TEM visualization [27].
Quantitative Morphological Assessment
For rigorous analysis, implement a systematic approach to quantify ultrastructural features:
Random Sampling: Examine multiple grid squares (minimum 10) at low magnification (2,000-5,000X) to ensure representative sampling.
Feature Scoring: Systematically document the presence of key morphological indicators (chromatin pattern, membrane integrity, organellar status) for each cell encountered.
Statistical Analysis: Calculate the percentage of cells displaying apoptotic versus necrotic features across multiple fields. A minimum of 100 cells per condition should be evaluated for statistical significance [21].
Advanced and Correlative Techniques
Complementary Methodologies for Cell Death Discrimination
While TEM provides the ultrastructural gold standard, several advanced techniques offer complementary approaches for distinguishing apoptosis from necrosis:
Table 2: Advanced Techniques for Cell Death Discrimination
| Technique | Principle | Advantages | Limitations |
|---|---|---|---|
| FF-OCT (Full-Field Optical Coherence Tomography) | Label-free interferometric imaging of cellular structural changes [15] | Non-invasive; enables real-time monitoring of dynamic processes; 3D surface topography mapping [28] | Lower resolution than TEM; limited subcellular detail |
| Capacitance Sensing | Measures changes in cell membrane capacitance during death processes [23] | Label-free; real-time monitoring; can distinguish apoptosis (monotonic decrease) from necrosis (step-like decreases) [23] | Does not provide visual morphological data; requires specialized equipment |
| Live-Cell FRET Imaging | Uses caspase-sensitive FRET probes with organelle-targeted fluorescent proteins [4] | Real-time discrimination at single-cell level; specific detection of caspase activation | Requires genetic engineering; potential phototoxicity during extended imaging |
Correlative Light Electron Microscopy (CLEM)
The integration of fluorescence microscopy with TEM through CLEM approaches provides a powerful strategy for linking dynamic molecular events with ultrastructural outcomes. This methodology allows researchers to first identify cells of interest using fluorescent markers (such as caspase sensors or viability dyes) before performing targeted ultrastructural analysis on the same cells, creating a direct correlation between biochemical and morphological data [27] [4].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Cell Death Research
| Reagent/Category | Specific Examples | Experimental Function |
|---|---|---|
| Apoptosis Inducers | Doxorubicin [15], Etoposide [22], TRAIL (TNF-related apoptosis-inducing ligand) [23] | Activate intrinsic or extrinsic apoptotic pathways in experimental models |
| Necrosis Inducers | High-concentration ethanol [15] [22], Hydrogen peroxide [22] [4], Freezing-thawing cycles [22] | Induce physicochemical damage leading to necrotic cell death |
| Molecular Detection Reagents | Annexin V conjugates [25], Propidium iodide [25] [24], TUNEL assay reagents [24], Caspase substrates/antibodies [4] | Detect biochemical markers of cell death pathways |
| TEM Reagents | Glutaraldehyde, Osmium tetroxide, Uranyl acetate, Epoxy resins [27] | Fix, contrast, and embed biological samples for ultrastructural analysis |
Technical Challenges and Methodological Considerations
Interpretation Pitfalls and Ambiguities
Researchers must remain cognizant of several challenges in ultrastructural analysis of cell death:
Temporal Dynamics: The transition from apoptosis to secondary necrosis represents a particular diagnostic challenge, as cells may display mixed morphological features [4]. Timely fixation and sampling are critical for accurate classification.
Cell-Type Specific Variations: Different cell types may manifest somewhat distinct morphological alterations during death processes. Establishing cell-type-specific baselines is essential [22].
Sample Preparation Artifacts: Improper fixation, processing, or sectioning can introduce artifacts that mimic pathological changes, such as membrane disruptions or organellar swelling [27]. Meticulous technique and appropriate controls are mandatory.
Emerging Techniques and Future Directions
Advanced EM techniques continue to evolve, offering new dimensions for cell death research. Cryo-electron microscopy preserves samples in a near-native state without chemical fixation, potentially revealing previously obscured ultrastructural details [27]. Volume EM approaches, including focused ion beam SEM and serial block-face imaging, enable three-dimensional reconstruction of entire cells or tissue volumes, providing unprecedented insight into spatial relationships during death processes [27].
Transmission electron microscopy remains an essential methodology for the definitive discrimination between apoptosis and necrosis, providing the ultrastructural resolution necessary to visualize the characteristic morphological signatures of each death pathway. While molecular techniques offer valuable complementary data, the detailed visualization of subcellular alterations afforded by TEM continues to serve as the gold standard in cell death classification. For researchers investigating early apoptosis, particularly in therapeutic contexts such as anticancer drug development, the integration of rigorous ultrastructural analysis with biochemical and live-cell approaches provides the most comprehensive framework for accurate cell death characterization.
Transmission Electron Microscopy (TEM) has played a foundational role in shaping our understanding of programmed cell death. The very conceptualization of apoptosis as a distinct form of cell death emerged not from biochemical or molecular techniques, but from meticulous ultrastructural observation using TEM. This technical guide explores the historical context of how TEM established the irreversible morphological criteria that continue to define apoptotic cells, providing an essential framework for researchers using TEM to identify early apoptosis in experimental and drug development settings.
The term apoptosis (from the Greek, meaning "to fall away from," as in leaves from a tree) was introduced in 1972 by Kerr, Wyllie, and Currie to describe a type of cell death previously referred to as "shrinkage necrosis" [29] [30]. Their observations, grounded in TEM analysis, established the characteristic ultrastructural features now considered the hallmark of apoptosis [29]. These features include cytoplasmic and nuclear condensation, nuclear fragmentation, normal morphological appearance of cytoplasmic organelles, and an intact plasma membrane [29]. Today, despite advances in biochemical and flow cytometric methodologies, TEM remains the gold standard for the specific identification of apoptotic cells based on these morphological criteria [29] [31].
The Historical Foundation: Kerr et al. (1972) and the Birth of Apoptosis
The seminal 1972 paper by Kerr, Wyllie, and Currie marked a paradigm shift in cell death research. Prior to their work, cell death was often loosely categorized, with terms like "necrobiosis," "shrinkage necrosis," or "chromatolysis" used inconsistently across different tissues and contexts [30]. The researchers recognized the need for an unambiguous term to describe a specific, regulated form of cell death observed in a wide variety of tissues, including during development and neoplastic transformation [29].
While apoptotic cells could sometimes be detected by light microscopy, it was the team's observations by transmission electron microscopy that provided the definitive evidence for establishing a new category of cell death [29]. TEM's superior resolving power revealed a conserved sequence of subcellular events that distinguished this process from accidental necrosis. The early TEM micrographs showed cells undergoing a controlled dismantling process, culminating in the formation of membrane-bound apoptotic bodies that were rapidly phagocytosed by neighboring cells without inciting an inflammatory response [29] [31]. This stood in stark contrast to the disruptive, inflammatory nature of necrotic cell death.
The Morphological Hallmarks of Apoptosis as Revealed by TEM
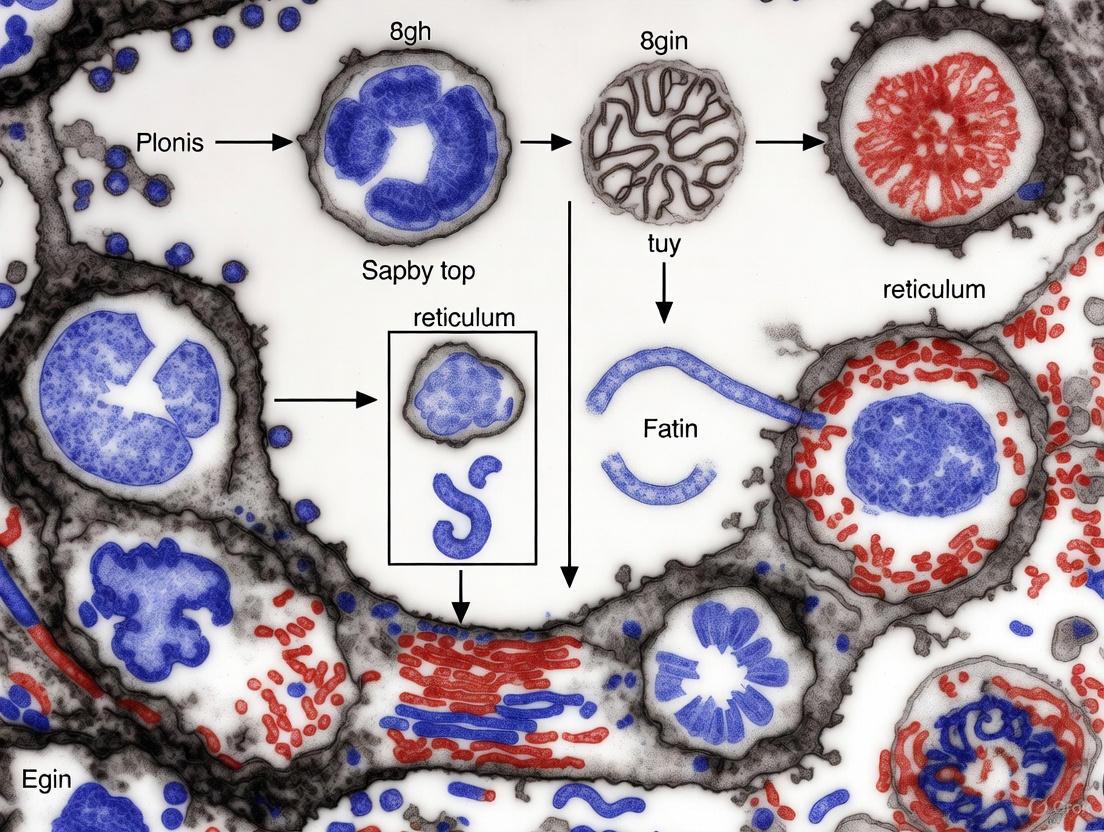
The criteria established via TEM form the basis for all modern apoptosis detection. The following table summarizes the key ultrastructural features that distinguish apoptosis from necrosis.
Table 1: Ultrastructural Criteria for Apoptosis vs. Necrosis as Defined by TEM
| Cellular Feature | Apoptosis | Necrosis |
|---|---|---|
| Cell Size | Condensation and shrinkage [3] [31] | Swelling (oncosis) [29] [3] |
| Plasma Membrane | Intact, but with blebbing and formation of apoptotic bodies [29] [32] | Ruptured and disrupted [29] [33] |
| Organelles | Generally normal morphology, though may be more tightly packed [29] | Swollen, especially mitochondria; disruption of membranes [29] |
| Nucleus | Chromatin condensation (pyknosis) and nuclear fragmentation (karyorrhexis); crescent-shaped masses at nuclear periphery [29] [3] [31] | Mild condensation followed by dissolution (karyolysis); no structured fragmentation [29] [3] |
| In Vivo Consequence | Phagocytosis by adjacent cells; no inflammation [29] [31] | Spilling of contents; associated inflammatory response [29] [33] |
Detailed Analysis of Apoptotic Morphology
Nuclear Changes: The most diagnostic feature of apoptosis is the fate of the nucleus. TEM reveals a characteristic progression: the chromatin first condenses into dense, coarse masses that marginate at the nuclear periphery, often assuming a striking crescent or 'half-moon' shape [29] [3]. This is followed by fragmentation of the entire nucleus into multiple, discrete, membrane-bound pyknotic bodies of condensed chromatin [29] [31] [32]. These changes are executed by endonucleases that cleave DNA at internucleosomal sites, but the morphological result is what is visualized by TEM [3].
Cytoplasmic and Membrane Events: Concurrent with nuclear collapse, the cell undergoes a reduction in volume (shrinkage) [3]. The cytoplasm becomes denser, although organelles like mitochondria typically retain their structural integrity initially [29]. A key feature is the preservation of the plasma membrane, which remains intact even as the cell surface blebs and eventually fragments into sealed, membrane-bound apoptotic bodies [29] [32]. These bodies contain variably condensed cytoplasm and nuclear fragments, and their formation prevents the leakage of immunogenic cellular contents, making apoptosis an immunologically "silent" process [29].
Experimental Protocols for TEM-Based Apoptosis Detection
Standard Sample Preparation for TEM Apoptosis Analysis
Reliable identification of apoptosis by TEM requires meticulous sample preparation to preserve ultrastructure.
- Protocol:
- Primary Fixation: Fix tissue samples or cell pellets immediately in a chilled glutaraldehyde solution (typically 2.5% in 0.1M sodium cacodylate buffer) for a minimum of 2 hours. This cross-links proteins and stabilizes cellular architecture [3].
- Washing: Rinse several times in the same buffer to remove excess fixative.
- Post-Fixation: Treat samples with a 1% osmium tetroxide solution for 1-2 hours. Osmium tetroxide acts as a secondary fixative and stains lipid membranes, enhancing contrast [3].
- Dehydration: Gradually dehydrate the samples using a graded series of ethanol (e.g., 50%, 70%, 90%, 100%) to prepare for resin infiltration.
- Infiltration and Embedding: Infiltrate the tissue with a resin, such as Epon or Spurr's, and then embed it in fresh resin for polymerization in an oven at 60°C for 24-48 hours [3].
- Sectioning and Staining: Use an ultramicrotome to cut ultrathin sections (60-90 nm). Mount sections on copper grids and stain with uranyl acetate and lead citrate to increase electron scattering and provide high-contrast images for the TEM [3].
Morphological Assessment and Quantification
- Imaging and Analysis: Systematically survey the prepared grids under the TEM. Identify apoptotic cells based on the consolidated criteria in Table 1. For quantification, capture images from multiple, randomly selected fields. The percentage of apoptotic cells can be calculated as (Number of apoptotic cells / Total number of cells) × 100. It is critical to assess multiple morphological features to confirm apoptosis, as some features can overlap with other death pathways like autophagy [3].
The Molecular Pathways of Apoptosis and Link to Morphology
The distinct morphology of apoptosis is the physical manifestation of a tightly regulated molecular cascade. TEM visualizes the endpoint of these signaling pathways, which are broadly classified as intrinsic and extrinsic.
Diagram 1: The intrinsic and extrinsic apoptotic pathways converge on effector caspases that execute the morphological changes visible by TEM.
The extrinsic pathway is triggered by the binding of extracellular death ligands (e.g., FasL) to cell surface death receptors. This leads to the recruitment of the adapter protein FADD and the activation of initiator caspase-8 [29]. The intrinsic pathway is initiated by internal cellular stresses (e.g., DNA damage), leading to the activation of BH3-only proteins, which promote the oligomerization of BAX and BAK in the mitochondrial outer membrane [29] [33]. This results in mitochondrial outer membrane permeabilization (MOMP) and the release of cytochrome c [33]. Cytochrome c then forms the apoptosome with APAF-1 and caspase-9, activating the executioner phase [29]. Both pathways converge on the activation of effector caspases (e.g., caspase-3), which cleave hundreds of cellular substrates, including those responsible for the structural breakdown of the nucleus (activation of CAD endonuclease) and cell (cleavage of cytoskeletal proteins), producing the characteristic morphology seen under TEM [29] [3] [33].
The Scientist's Toolkit: Key Reagents for Apoptosis Research
Table 2: Essential Research Reagents for Apoptosis Detection and Analysis
| Reagent / Assay | Function and Application in Apoptosis Research |
|---|---|
| Glutaraldehyde & Osmium Tetroxide | Primary and secondary fixatives for TEM sample preparation; preserve ultrastructure and enhance membrane contrast [3]. |
| Uranyl Acetate & Lead Citrate | Heavy metal stains for TEM grids; bind to cellular components to increase electron scattering and image contrast [3]. |
| Hoechst 33342 / DAPI | Fluorescent DNA-binding dyes used in fluorescence microscopy to visualize nuclear condensation and fragmentation, hallmarks of apoptosis [3] [32]. |
| Annexin V-FITC/PI | Flow cytometry/fluorescence microscopy assay to detect phosphatidylserine (PS) externalization (early apoptosis) and loss of membrane integrity (late apoptosis/necrosis) [32]. |
| JC-1 Dye | Lipophilic cationic fluorescent dye used to measure mitochondrial membrane potential (ΔΨm); loss of potential is an early indicator of intrinsic apoptosis [32]. |
| TUNEL Assay | Detects DNA fragmentation (a late apoptotic event) by labeling 3'-OH ends of broken DNA strands; can be used in tissue sections or cells [31] [32]. |
| Antibodies to Cleaved Caspase-3 & PARP | Western blot or immunohistochemistry markers for biochemical confirmation of apoptosis execution; detect specific proteolytic cleavage events [32]. |
Transmission Electron Microscopy provided the foundational context for our understanding of apoptosis. The morphological criteria established by Kerr et al. in 1972—cell shrinkage, chromatin condensation and margination, nuclear fragmentation, and formation of intact apoptotic bodies—remain the definitive characteristics of this programmed cell death process [29] [31]. For researchers and drug development professionals, TEM continues to offer the most direct and unambiguous method for identifying apoptotic cells and distinguishing them from those dying by other mechanisms. While biochemical and flow cytometric assays provide valuable, often quantitative data on apoptotic pathways, they should be interpreted in conjunction with morphological assessment by light or electron microscopy to confirm the specific mode of cell death [29] [3]. Thus, TEM remains an indispensable tool in the cell biologist's arsenal, providing the ultrastructural gold standard against which newer methods are often validated.
From Cell to Image: A Methodological Protocol for TEM-Based Apoptosis Detection
The precise ultrastructural identification of early apoptosis via transmission electron microscopy (TEM) is critically dependent on the quality of sample preparation. suboptimal fixation, dehydration, or embedding can obscure key morphological hallmarks, leading to misclassification of cell death modalities. This technical guide provides a comprehensive, evidence-based framework for the preparation of apoptotic cell samples, detailing protocols designed to preserve the subtle cellular changes characteristic of early apoptosis. Within the broader context of TEM-based apoptosis research, we emphasize strategies to balance ultrastructural preservation with antigenicity retention, incorporate quantitative analysis, and avoid common artifacts that compromise data interpretation for researchers and drug development professionals.
Transmission electron microscopy (TEM) remains the gold standard for the definitive identification of early apoptosis, enabling the visualization of key morphological hallmarks such as chromatin condensation, cellular shrinkage, and membrane blebbing at the nanoscale [14]. However, the fidelity of this ultrastructural analysis is entirely contingent upon the quality of sample preparation. Inadequate fixation, dehydration, and embedding can distort or obliterate these delicate features, leading to the misclassification of cell death modalities—a significant concern in both basic research and preclinical drug development [34] [14]. This whitepaper provides an in-depth guide to optimized protocols for the preparation of apoptotic cells for TEM, framed within the rigorous requirements of academic and industrial microscopy research. The procedures outlined herein are designed to achieve the critical balance between optimal preservation of cellular morphology and the retention of biomolecular antigenicity for potential immunogold labeling, ensuring that researchers can capture a definitive and unambiguous snapshot of early apoptotic events.
Fundamental Principles of Apoptosis for TEM Analysis
A thorough understanding of apoptotic morphology is a prerequisite for meaningful TEM analysis. The following hallmarks are primary diagnostic targets that sample preparation must faithfully preserve.
- Early Apoptotic Events: The initial stages are characterized by chromatin condensation (margination against the nuclear envelope), cell shrinkage, and loss of cell-cell contacts. The cytoplasm may become denser, and organelles like mitochondria often remain structurally intact initially [14].
- Mid to Late Apoptotic Events: The process advances to nuclear fragmentation (karyorrhexis) and budding of the cell to form membrane-bound apoptotic bodies. These bodies contain tightly packed cytoplasm and nuclear fragments, and the plasma membrane remains intact, preventing the release of pro-inflammatory cellular contents [14].
- Contrast with Other Cell Death Modalities: Accurate diagnosis requires distinguishing apoptosis from other processes. Oncosis (a prelude to necrosis) features cellular and organellar swelling, not shrinkage [34]. Pyroptosis involves plasma membrane pore formation and inflammatory cytokine release, while necroptosis shares some apoptotic features but ultimately results in membrane rupture [14]. Proper sample preparation is paramount for making these critical distinctions, as techniques like flow cytometry using annexin V/PI can misclassify oncotic cells as apoptotic due to phosphatidylserine exposure [34].
Core Technical Protocols
Fixation Strategies for Apoptotic Cells
Fixation is the most critical step, as it terminates biochemical activity and stabilizes cellular structures in their native state. The goal is to rapidly immobilize lipids and proteins without introducing artifacts or masking antigenic epitopes.
Table 1: Key Fixatives for Apoptosis TEM Studies
| Fixative | Penetration Ability | Primary Effect | Impact on Apoptotic Morphology | Recommendation for Apoptosis Studies |
|---|---|---|---|---|
| Glutaraldehyde | Strong | Crosslinks proteins; excellent structural preservation | Can cause tissue shrinkage; may mask antigen epitopes | Essential for core structural integrity; use at low concentrations (0.5-2%) in combination with PFA. |
| Paraformaldehyde (PFA) | Stronger than Glutaraldehyde | Crosslinks proteins; faster penetration | Better preservation of antigen activity | Use as a primary fixative (2-4%) in a mixture with low-concentration glutaraldehyde. |
| Osmium Tetroxide | Mild | Stabilizes lipids; adds electron density | Severely destroys antigen activity; can swell tissue | Use as a post-fixative after aldehyde fixation to preserve membrane structures. Avoid if IEM is planned. |
| Tannic Acid | Mild | Enhances contrast of membranes and proteins | Can mask epitopes and increase background | Use as an additive to primary fixative to improve membrane visualization. |
| Glyoxal | Strong | Crosslinks membrane and cytoskeletal proteins | Low pH may enhance epitope exposure differences | An emerging alternative for improved antigen preservation under mild conditions [35]. |
A recommended primary fixation protocol for apoptotic cells in culture is as follows:
- Prepare Fixative Cocktail: Mix 2-4% paraformaldehyde with 0.5-2% glutaraldehyde in a 0.1 M phosphate or cacodylate buffer (pH 7.4). The buffer must contain sucrose to maintain osmolarity.
- Apply Fixative: For adherent cells, gently remove the culture medium and immediately add the pre-warmed (37°C) fixative. For suspension cells, pellet the cells gently and resuspend in the fixative. Note: Protocol adjustments for adherent cells are critical, as including all cells in the medium can lead to overestimation of cell death by incorporating already-dead cells into the analysis [36].
- Fixation Duration: Fix at room temperature for 1-2 hours, followed by storage at 4°C if necessary. Prolonged fixation in high-concentration glutaraldehyde should be avoided to prevent excessive cross-linking.
Dehydration and Embedding
Following thorough buffer washing to remove excess fixatives, samples must be dehydrated and embedded in a resin that permits ultrathin sectioning.
- Dehydration: A graded series of ethanol or acetone is used to remove water from the fixed specimen. A typical series is: 30% → 50% → 70% → 90% → 100% → 100% ethanol, with 10-15 minutes per step. The final 100% ethanol step should be performed with anhydrous ethanol to ensure complete dehydration.
- Resin Infiltration and Embedding: The dehydrated sample is infiltrated with a resin mixture that later hardens. For optimal preservation of antigenicity, low-temperature embedding resins are recommended.
- Epoxy Resins (Epon, Araldite): Provide excellent ultrastructural preservation and sectioning properties but require high-temperature polymerization (~60°C) that can destroy most antigenic sites. Best used for morphological studies without immunolabeling.
- Acrylic Resins (LR White, Lowicryl): These can be polymerized at low temperatures (-35°C to 4°C) via UV light. This process significantly better preserves protein antigenicity, making them the resins of choice for correlative immunoelectron microscopy (IEM) studies of apoptosis biomarkers [35]. The infiltration involves a gradual transition from ethanol to resin (e.g., 1:2, 1:1, 2:1 resin:ethanol, then pure resin) before final polymerization in embedding molds.
The following diagram illustrates the complete workflow for sample preparation, integrating the key decision points for apoptosis research.
Immunoelectron Microscopy (IEM) for Apoptosis
IEM combines ultrastructural imaging with the molecular specificity of immunolabeling, allowing for the precise localization of apoptosis-related proteins (e.g., caspases, Bax) within the cell. The choice of IEM strategy depends heavily on the sample preparation steps above [35].
- Pre-embedding Labeling: Antibodies are applied to the sample before resin embedding. This method offers better access to epitopes, especially for low-abundance antigens, but requires permeabilization that can damage ultrastructure.
- Post-embedding Labeling: This is the most common approach. Labeling is performed on the surface of ultrathin sections after embedding. It provides superior structural preservation but epitopes may be masked by the resin. Using low-temperature acrylic resins (e.g., LR White) is crucial for success in post-embedding IEM [35].
- Tokuyasu Method: A powerful cryotechnique where samples are lightly fixed, infused with sucrose, frozen, and sectioned at low temperatures. The frozen sections are then immunolabeled. This method offers an exceptional balance between antigen preservation and structural integrity and is highly suited for apoptosis biomarker research [35].
Advanced and Integrated Methodologies
To enhance the rigor and depth of TEM-based apoptosis research, several advanced methodologies can be integrated.
- Quantitative Ultrastructural Analysis: Moving beyond qualitative description is key. A framework based on systematic uniform random sampling (SUR) ensures unbiased data collection. Furthermore, deep learning algorithms (e.g., Gold Digger for automated quantification of immunogold particles) and 3D reconstruction via FIB-SEM (with isotropic resolution reaching ~5 nm) enable robust statistical analysis of apoptotic events and biomarker distribution [35].
- Correlative Light and Electron Microscopy (CLEM): CLEM is a transformative multimodal integration strategy. It allows researchers to first visualize dynamic processes like caspase activation in live cells using fluorescent probes (e.g., FLICA probes) and then relocate the very same cell for high-resolution TEM analysis. This directly links functional data to structural context, providing an unparalleled view of the apoptotic cascade [35] [37].
- Pathway-Specific Pharmacological Inhibition: To confirm the apoptotic nature of observed cell death, researchers can use specific inhibitors during cell culture. For example, the pan-caspase inhibitor Z-VAD-FMK can be applied to cells prior to an apoptotic stimulus. A significant reduction in cells displaying apoptotic morphology by TEM provides strong mechanistic evidence for caspase-dependent apoptosis, helping to distinguish it from other death pathways like pyroptosis [38].
The following diagram maps the core apoptotic signaling pathways and their connection to the ultrastructural hallmarks visible via TEM.
The Scientist's Toolkit: Essential Reagents and Materials
Table 2: Key Research Reagent Solutions for Apoptosis TEM Preparation
| Reagent/Material | Function/Application | Key Considerations |
|---|---|---|
| Glutaraldehyde (EM Grade) | Primary fixative for cross-linking proteins; provides superb structural integrity. | Use at low concentrations (0.5-2%) in combination with PFA to minimize antigen masking and tissue shrinkage [35]. |
| Paraformaldehyde (EM Grade) | Primary fixative; rapidly penetrates cells to stabilize proteins. | Typically used at 2-4%. A mixture with glutaraldehyde offers a balance of speed and strength of fixation [35]. |
| Osmium Tetroxide | Post-fixative; stabilizes phospholipids and adds electron density to membranes. | Destroys antigenicity; use only for morphological studies. Requires careful handling and disposal [35]. |
| LR White Resin | Low-temperature acrylic embedding medium. | Polymerized with UV light at low temps; superior for preserving antigenicity for post-embedding IEM [35]. |
| Lowicryl Resins (K4M/HM20) | Low-temperature embedding resins. | Polymerized at -35°C; ideal for preserving ultrastructure and antigen activity for sensitive IEM applications [35]. |
| Z-VAD-FMK | Cell-permeable, irreversible pan-caspase inhibitor. | Used as a functional tool (at 10-100 μM) to confirm caspase-dependence of observed cell death, distinguishing apoptosis from other pathways [38]. |
| Colloidal Gold Conjugates | Electron-dense markers for immunolabeling in IEM. | Particle size (e.g., 5-30 nm) can be tuned for different labeling densities and resolution requirements [35]. |
The reliable identification of early apoptosis by TEM is a cornerstone of cell death research, with direct implications for understanding fundamental biology and evaluating the mechanisms of action of novel therapeutics. This guide has delineated a comprehensive pathway from sample fixation to final embedding, underscoring the necessity of methodical optimization at each stage. By adhering to these detailed protocols for fixation cocktails, dehydration series, and resin selection—and by integrating advanced quantitative and correlative techniques—researchers can achieve an unparalleled preservation of the fleeting ultrastructural signatures of apoptosis. The resulting high-fidelity data is essential for building robust, conclusive models of cellular demise in health and disease.
This technical guide details the application of uranyl acetate and lead citrate staining for transmission electron microscopy (TEM) in the context of apoptosis research. The "double contrasting" technique is a standard method for enhancing the visibility of subcellular structures, crucial for identifying early apoptotic events such as membrane blebbing, chromatin condensation, and organelle alteration. This whitepaper provides in-depth protocols, safety considerations, and data interpretation guidelines to support researchers in drug development in leveraging ultrastructural analysis for evaluating novel anticancer therapeutics.
In transmission electron microscopy, the inherent contrast of biological specimens is low because cellular components comprise elements of similar atomic numbers. Staining with heavy metals is therefore essential to create sufficient electron density differences for high-resolution imaging. Uranyl acetate (UA) and lead citrate represent the cornerstone of this process, a combination often called "double contrasting" [39]. When used sequentially, these stains provide comprehensive contrast enhancement across various cellular structures, making them indispensable for morphological studies.
The analysis of early apoptosis presents a prime application for this technique. Apoptosis is a tightly regulated process characterized by specific morphological changes, including cell shrinkage, chromatin condensation, and plasma membrane blebbing. Distinguishing these features from necrosis or other cellular states requires the high-resolution capability that only well-stained TEM samples can provide. Furthermore, with the rise of nanoparticle-based cancer therapies designed to induce apoptosis, such as those utilizing magnetic iron oxide or hybrid silver-iron oxide nanoparticles, verifying the mechanism of action at the ultrastructural level has become a critical step in preclinical drug development [40] [41].
Scientific Basis of Staining
Mechanism of Action
Heavy metal stains interact with biological tissues by binding to specific macromolecular components. The resulting variation in electron scattering power creates the contrast observed in TEM images.
Uranyl Acetate (UA): Uranyl ions (UO₂²⁺) exhibit a high atomic weight (238) and act as a cationic stain that binds preferentially to negatively charged groups in tissues [39]. These include:
- Phosphate groups on nucleic acids (DNA and RNA).
- Carboxyl groups in proteins and sialic acid residues of glycoproteins and gangliosides.
- This binding profile makes UA exceptionally good for contrasting membranes, nucleic acids, and ribosomes [39].
Lead Citrate: Lead ions (Pb²⁺) function as a general stain at high pH (approximately pH 12). Its contrasting effect is enhanced by prior treatment with reduced osmium and UA [39]. It primarily interacts with:
- Protein components throughout the cytoplasm.
- Hydroxyl groups in carbohydrates like glycogen.
- Lead citrate provides robust staining of ribosomes, lipid membranes, the cytoskeleton, and other cytoplasmic compartments [39].
Table 1: Staining Affinities of Uranyl Acetate and Lead Citrate
| Cellular Structure | Uranyl Acetate Affinity | Lead Citrate Affinity | Primary Staining Mechanism |
|---|---|---|---|
| Plasma Membrane | High | High | UA: Binds lipid head groups; Lead: Interacts with osmium deposits |
| Nuclear Chromatin | High (via DNA) | Moderate | UA: Binds DNA phosphate backbone |
| Mitochondria | Moderate | High | Lead: Stains membranes & matrix proteins |
| Ribosomes | High (via RNA) | High | UA: Binds RNA; Lead: Binds proteins |
| Endoplasmic Reticulum | Moderate | High | Lead: Stains membranous and protein content |
| Glycogen Granules | Low | High | Lead: Binds hydroxyl groups |
The Double Staining Workflow
The sequential application of UA followed by lead citrate is critical for achieving optimal results. Uranyl acetate acts as a "mordant," or a substance that fixes a dye, by binding to tissues and creating sites that enhance the subsequent deposition of lead. This synergistic interaction produces a broader and more intense contrast than either stain used alone [39]. The following diagram illustrates the sequential staining process and its mechanistic basis.
Technical Protocols
Reagent Preparation
Proper preparation of staining solutions is fundamental to success and directly impacts staining quality and the avoidance of artifacts.
Table 2: Research Reagent Solutions for TEM Staining
| Reagent | Composition / Function | Critical Handling Notes |
|---|---|---|
| Aqueous Uranyl Acetate | 0.5% - 4% (w/v) in distilled water; contrasts nucleic acids and membranes [39]. | Light-sensitive; store in brown bottle at 4°C. Highly toxic and mildly radioactive; use full protective gear. |
| Lead Citrate (Reynolds) | 3% solution from lead nitrate and sodium citrate; stains proteins and carbohydrates [39] [42]. | Extremely toxic. Prepare with CO₂-free water and under protective atmosphere (e.g., helium) to prevent lead carbonate precipitate. |
| Sodium Hydroxide | Pellets or concentrated solution; used to create alkaline environment for lead citrate solubility and to avoid precipitate [43]. | Caustic; handle with care. |
| CO₂-free Water | Double-distilled, boiled, or degassed water; essential for preparing stable lead citrate solution without precipitation [39]. | Prepare fresh or store sealed to prevent CO₂ absorption. |
| 0.25% Oxalic Acid | Filtered solution; used to remove uranyl acetate and lead salt precipitates from sections if necessary [43]. | Use with caution on delicate resin sections. |
Materials: Latex gloves, lab coat, protective mask, 200 ml brown stock bottle, 100 ml volumetric flask, balance under fume hood, weighing dish, magnetic stirrer, hot plate, Whatman #1 filter paper, glass funnel, uranyl acetate dihydrate powder (depleted), CO₂-free double-distilled water. Procedure:
- Weigh 4 g of uranyl acetate under a fume hood with full protective equipment.
- Add the powder to a 100 ml volumetric flask.
- Pipette 96 ml of near-boiling CO₂-free double-distilled water into the flask.
- Place on a stirrer until the crystals are completely dissolved. Allow the solution to cool to room temperature.
- Filter the solution through Whatman #1 filter paper into the 200 ml brown bottle.
- Cap tightly, label clearly, and store at 4°C. The solution is typically stable for several months.
Materials: Lead nitrate, tri-sodium citrate dihydrate, 1M sodium hydroxide, CO₂-free double-distilled water, volumetric flask, stirrer. Procedure:
- Dissolve 1.33 g of lead nitrate and 1.76 g of tri-sodium citrate in 30 ml of CO₂-free distilled water in a 50 ml flask.
- Shake the mixture vigorously for 1 minute. A milky white suspension will form.
- Allow the mixture to stand for 30 minutes with intermittent shaking. The citrate should dissolve.
- Add 8.0 ml of 1M sodium hydroxide to the clear solution. The mixture will turn clear as lead citrate forms.
- Dilute the solution to 50 ml with CO₂-free distilled water.
- Store the solution in an airtight, CO₂-free environment (e.g., a syringe or bottle with minimal headspace) to prevent formation of insoluble lead carbonate.
Staining Procedure for Ultrathin Sections
The following protocol describes the manual staining of grids bearing ultrathin sections.
Materials:
- Grids with ultrathin sections (50-90 nm)
- Prepared solutions of uranyl acetate and lead citrate
- Parafilm
- Forceps
- Two beakers of warm distilled water (approx. 50°C) [43]
- One beaker of 0.1M sodium hydroxide [43]
- Waste disposal for heavy metals
Detailed Protocol:
- Preparation: Place droplets of uranyl acetate and lead citrate solutions on clean strips of Parafilm. Place a small pellet of sodium hydroxide near the lead citrate droplet to absorb atmospheric CO₂ [43] [39].
- Uranyl Acetate Staining:
- Using fine forceps, float the grid (section side down) on a droplet of uranyl acetate.
- Stain for 2 to 10 minutes at room temperature, protected from light.
- Rinsing:
- Rinse the grid thoroughly by dipping it sequentially in two beakers of warm distilled water. The warm water helps prevent the formation of uranyl acetate precipitates [43].
- Carefully blot the grid edge on filter paper to remove excess water.
- Lead Citrate Staining:
- Float the grid on the lead citrate droplet.
- Stain for 1 to 5 minutes at room temperature.
- Final Rinsing:
- Rinse the grid by dipping it first in the 0.1M sodium hydroxide solution, then thoroughly in two beakers of warm distilled water. The alkaline dip neutralizes any residual CO₂ that could cause lead carbonate precipitation.
- Drying and Storage: Blot the grid dry and store in a grid box for TEM observation.
Troubleshooting: Artifact Prevention and Removal
Precipitates are the most common artifacts encountered with these stains.
- Lead Carbonate Precipitate: Appears as fine black grains or a fine deposit on sections [39]. Prevention: Use CO₂-free water for all solutions and rinses, protect lead citrate from air exposure with NaOH pellets or an inert atmosphere [43] [39].
- Uranyl Acetate Precipitate: Appears as yellow, needle-like crystals [39]. Prevention: Use warm distilled water for rinsing, filter the stain regularly, and protect it from light.
- Remediation: If precipitates form on sections, they can sometimes be removed by gently dipping the grid in a weak acid like 0.25% filtered oxalic acid for a duration 3-4 times longer than the original stain time [43]. This must be done cautiously, as it can damage the section.
Application in Apoptosis Research
The precise morphological detailing enabled by UA/lead citrate staining is vital for confirming apoptosis induction by novel therapeutics, a key area in drug development.
Identifying Key Apoptotic Features
TEM allows researchers to distinguish early apoptosis from other forms of cell death based on ultrastructural criteria:
- Early Apoptosis: Chromatin condenses into dense masses that marginate against the nuclear envelope. The cell and nucleus shrink, but membrane integrity is maintained. Uranyl acetate vividly highlights the condensed chromatin, while both stains delineate the intact but convoluted plasma membrane [40].
- Late Apoptosis: The nucleus fragments, and the cell blebs into apoptotic bodies. These membrane-bound bodies, often containing cytoplasmic elements or nuclear fragments, are clearly defined by the staining.
- Necrosis: Characterized by swelling of organelles and rupture of the plasma membrane, presenting a starkly different stained appearance from apoptosis.
Case Study: Validating Nanoparticle-Induced Apoptosis
Research on magnetic iron oxide nanoparticles (MIONS) coated with bacterial exopolysaccharide (EPS) for breast cancer treatment exemplifies this application. In vitro studies on MCF-7 breast cancer cells demonstrated that EPS/MIONS significantly reduced cell viability. Staining and flow cytometry analysis revealed that the percentage of early apoptotic cells was as high as 71.87% for EPS/MIONS-treated groups compared to controls [40]. While flow cytometry quantifies apoptosis, TEM with UA/lead citrate staining is required to visually confirm the classic morphology, thereby validating the mechanism of action and ruling out necrotic death.
Similar approaches are used in developing therapies for other cancers, such as glioblastoma, where hybrid silver-iron oxide nanoparticles (SIONFs) are investigated for their theranostic potential and ability to induce apoptosis [41]. The following workflow diagrams the integration of staining within a therapeutic efficacy study.
Advanced Techniques and Future Directions
While post-section staining is routine, en bloc staining—staining the tissue block before sectioning—is gaining traction for advanced EM techniques like serial block-face SEM (SBF-SEM) and array tomography. This method involves treating the sample with combinations of heavy metals (e.g., UA, lead aspartate, osmium-thiocarbohydrazide-osmium (OTO)) during processing. It provides superior contrast for imaging large tissue volumes and minimizes handling of individual grids, though it requires careful optimization to ensure even stain penetration [44] [45].
The field continues to evolve with the development of automated staining systems, such as the Leica EM AC20, which standardize the process, minimize user contact with hazardous reagents, and virtually eliminate problems with precipitation, thereby enhancing reproducibility and safety [39].
The double-staining technique using uranyl acetate and lead citrate remains a fundamental and powerful tool in the arsenal of cell biology and drug development. Its ability to provide high-contrast, high-resolution images of subcellular morphology is unparalleled for the definitive identification of apoptotic events. Mastering the protocols, understanding the mechanisms, and diligently applying safety measures allows researchers to reliably generate quality data that can critically support the evaluation of new anticancer agents, from recombinant proteins like TRAIL to advanced nanoparticle systems.
In the context of transmission electron microscopy (TEM) research for identifying early apoptosis, capturing diagnostic ultrastructural features at high magnification is paramount. Apoptosis, or programmed cell death, was originally defined based solely on morphological criteria observable by TEM, establishing this technique as the historical and continuing "gold standard" for its specific identification [29]. In early apoptosis, morphological reorganization occurs before conventional cellular markers become detectable, preceding active phases of the apoptotic process [46]. This technical guide details the methodologies for using high-resolution imaging to capture these subtle yet critical early ultrastructural changes, providing researchers and drug development professionals with precise protocols for their investigative work.
Core Ultrastructural Features of Early Apoptosis
The following table summarizes the key diagnostic ultrastructural features of early apoptosis identifiable via TEM, based on the original morphological criteria established by Kerr et al. [29].
Table 1: Diagnostic Ultrastructural Features of Early Apoptosis
| Cellular Component | Early Apoptotic Morphology | Significance |
|---|---|---|
| Nucleus | Chromatin condensation (pyknosis) and marginalion against the nuclear envelope; nuclear fragmentation (karyorrhexis) [29]. | Primary hallmark of apoptosis; precedes other changes [46]. |
| Plasma Membrane | Loss of microvilli and cell shrinkage; membrane blistering [47]. | Indicates early loss of cytoplasmic volume. |
| Mitochondria | Pre-apoptotic alterations, but normal morphological appearance is generally maintained initially [29]. | Early event in intrinsic pathway; can occur before nuclear changes [47]. |
| Perinuclear Membrane | May show changes in structural complexity, becoming less rough [46]. | Reflects overall loss of structural integrity. |
| Overall Cell Structure | Formation of membrane-bound apoptotic bodies containing condensed cytoplasm and organelles [29]. | Final packaging of the cell for phagocytosis. |
Experimental Protocols for TEM Analysis
Sample Preparation for TEM Ultrastructural Analysis
A critical protocol for identifying early apoptosis involves processing cells for TEM observation. The following workflow, adapted from studies on SK-BR-3 breast cancer cells and in vivo models, ensures preservation of delicate ultrastructural features [48] [46] [47].
- Fixation: Immediately after treatment or isolation, fix cell pellets or tissue samples (≤1 mm³) in a mixture of 3% paraformaldehyde and 1% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for a minimum of 48 hours at 4°C. This cross-linking fixative is essential for preserving subcellular structures.
- Post-Fixation: Rinse samples in buffer and post-fix with 1% osmium tetroxide for 1-2 hours. Osmium tetroxide stabilizes lipids and provides electron density to membranes.
- Dehydration: Gradually dehydrate the samples using an ascending ethanol series (e.g., 50%, 70%, 90%, 100%).
- Embedding: Infiltrate and embed the tissue in a hard epoxy resin, such as Epon 812.
- Sectioning: Using an ultramicrotome, cut ultrathin sections (typically 50-90 nm thick) and mount them on copper or nickel grids.
- Staining: For contrast, stain the sections with heavy metals: first with uranyl acetate (aqueous or alcoholic), then with lead citrate.
In Vivo Model for Therapy Monitoring
To study the induction of apoptosis by a therapeutic agent in vivo, the following methodology can be employed, as demonstrated in studies of toxin T-514 and trastuzumab [48] [47].
- Animal Model: Use an immunocompromised mouse model (e.g., BALB/c nude mice).
- Xenograft Implantation: Implant human cancer cells (e.g., SK-BR-3 for HER2-positive breast cancer) into the mammary fat pads.
- Treatment: Once tumors reach a target volume (e.g., ~140 mm³), administer the therapeutic agent (e.g., 20 mg/kg trastuzumab via intraperitoneal injection). Control groups receive a vehicle solution.
- Tissue Harvest: At predetermined time points post-treatment (e.g., 4, 8, 12, 24 hours), euthanize the animals and harvest the target tissues (e.g., tumor, kidney, lung).
- Analysis: Process the tissues for histopathology, TEM ultrastructural analysis, and biochemical assays (e.g., DNA fragmentation) to correlate morphology with biochemical evidence of apoptosis.
Figure 1: TEM sample preparation workflow for apoptosis detection.
Quantitative Ultrastructural Analysis
Beyond qualitative observation, TEM allows for quantitative morphometric analysis to objectively characterize early apoptotic changes. Fractal dimension (FD) analysis is one such method that measures the structural complexity of cellular components [46].
Table 2: Fractal Dimension (FD) Analysis of Ultrastructure in Early Apoptosis
| Cellular Structure | FD in Control Cells | FD in Early Apoptosis | Interpretation |
|---|---|---|---|
| Nuclear Chromatin | Higher FD value | Reduced FD value [46] | Loss of structural complexity and textural reorganization. |
| Plasma Membrane | Higher FD value | Lower FD value [46] | Membrane becomes less rough and complex. |
| Perinuclear Membrane | Unchanged FD value | Unchanged FD value (initially) [46] | Not an early marker in all cell types. |
This quantitative approach confirms that morphological reorganization, detectable by a loss of structural complexity (reduced FD), occurs in the early stages of apoptosis, often before other conventional markers are evident [46].
Correlative and Functional Assays
While TEM provides unmatched ultrastructural detail, correlating these findings with biochemical and functional assays strengthens the identification of apoptosis. Key methodologies include:
- DNA Fragmentation Analysis: A hallmark biochemical event in apoptosis is internucleosomal DNA cleavage. This can be visualized using gel electrophoresis to detect a "DNA ladder" pattern and quantified spectrophotometrically. Studies show fragmented DNA levels can vary by tissue, for example, being highest in the liver, followed by the kidney and lung in toxin-induced apoptosis [47].
- TUNEL Assay (Terminal deoxynucleotidyl transferase dUTP Nick End Labeling): This technique enzymatically labels the 3'-ends of DNA fragments in situ, allowing for the light-microscopic visualization of cells with degraded DNA. TUNEL-positive cells can be detected as early as 4 hours post-intoxication in in vivo models [47]. It is crucial to interpret TUNEL results alongside morphology, as this technique can also stain necrotic cells [29].
Figure 2: Key morphological & biochemical events in apoptosis.
The Scientist's Toolkit: Research Reagent Solutions
The following table details essential materials and reagents used in the preparation and analysis of samples for the ultrastructural identification of early apoptosis.
Table 3: Essential Research Reagents for TEM Apoptosis Analysis
| Reagent / Material | Function / Application |
|---|---|
| Glutaraldehyde / Paraformaldehyde | Primary fixatives that cross-link proteins and preserve cellular ultrastructure for TEM [48] [47]. |
| Osmium Tetroxide | Post-fixative that stabilizes lipids and imparts electron density to membranes, enhancing contrast [48]. |
| Epon 812 Epoxy Resin | Embedding medium that provides firm support for cutting ultrathin sections with an ultramicrotome [48] [46]. |
| Uranyl Acetate & Lead Citrate | Heavy metal stains that bind to cellular components (e.g., nucleic acids, membranes), providing contrast in TEM imaging [29]. |
| Trastuzumab (Herceptin) | Monoclonal antibody therapeutic used in research to induce apoptosis in HER2-positive breast cancer xenograft models [48]. |
| SK-BR-3 Cell Line | A human breast cancer cell line that overexpresses HER2, commonly used in apoptosis research related to targeted therapy [48] [46]. |
| Annexin V | A protein that binds to phosphatidylserine (PS) exposed on the outer leaflet of the plasma membrane, a marker for early apoptosis detectable via fluorescence microscopy or flow cytometry [48] [49]. |
The precise capture of diagnostic ultrastructural features via transmission electron microscopy remains a cornerstone of early apoptosis research. By employing rigorous sample preparation protocols, correlating ultrastructural findings with biochemical assays, and utilizing quantitative methods like fractal morphometry, researchers can reliably identify the earliest stages of programmed cell death. This detailed morphological analysis is indispensable for validating the efficacy of novel therapeutic agents and advancing our understanding of cell death mechanisms in drug development.
Within the framework of transmission electron microscopy (TEM) research for identifying early apoptosis, systematic image analysis provides a powerful, quantitative approach to complement traditional ultrastructural assessment. While TEM remains the gold standard for visualizing the distinctive morphological hallmarks of programmed cell death, integrating standardized image analysis protocols enables researchers to transform qualitative observations into objective, reproducible data. This technical guide provides researchers, scientists, and drug development professionals with a structured checklist and methodological framework for identifying early apoptotic indicators through image analysis, ensuring precision and reliability in cellular death characterization.
Ultrastructural Hallmarks of Early Apoptosis: A Quantitative Framework
The definitive identification of early apoptosis via TEM relies on recognizing specific subcellular changes that precede complete cellular disintegration. Systematic analysis of these features allows for distinction from other forms of programmed cell death.
Table 1: Quantitative Ultrastructural Indicators of Early Apoptosis via TEM
| Cellular Feature | Early Apoptotic Morphology | Alternative PCD Morphology (for contrast) | Key Distinguishing TEM Characteristics |
|---|---|---|---|
| Nucleus | Chromatin condensation (pyknosis), margination along nuclear envelope | Peripheral chromatin condensation (Necroptosis); Nuclear dilation (Paraptosis) | Electron-dense, granular chromatin aggregates; intact nuclear envelope initially [26] [50] |
| Cytoplasm & Organelles | Organelles generally intact; cytoplasmic compaction | Massive vacuolization (Autophagy); Organelle swelling (Necroptosis) | Preservation of mitochondrial cristae; dilated endoplasmic reticulum; condensed cytoplasmic matrix [26] [14] |
| Cell Membrane | Preservation of membrane integrity; budding | Rapid plasma membrane rupture (Pyroptosis/Necroptosis) | Intact plasma membrane with blebbing; exposure of phosphatidylserine [50] |
| Mitochondria | Normal ultrastructure or condensed morphology | Swelling & rupture (Necroptosis); shrinkage (Ferroptosis) | No mitochondrial swelling; release of Cytochrome c without ultrastructural collapse [14] [50] |
The primary morphological changes of early apoptosis, as visualized by TEM, include chromatin condensation into dense, marginated masses and cellular shrinkage with compaction of cytoplasmic constituents. A critical diagnostic indicator is the preservation of organelle integrity, particularly in mitochondria, which distinguishes it from necrotic processes. Furthermore, the cell membrane remains intact but exhibits zeiosis (blebbing), eventually forming membrane-enclosed apoptotic bodies containing fragmented nuclei and organelles [24] [26] [50].
Analytical Workflow for Apoptosis Detection
A robust, multi-technique approach is recommended to confirm early apoptosis, correlating TEM's morphological gold standard with biochemical and molecular techniques.
Diagram 1: Multi-modal apoptosis detection workflow (76 characters)
Detailed TEM Protocol for Apoptosis Detection
This protocol is adapted from established methodologies for ultrastructural analysis of cell death [51] [26].
Sample Preparation:
- Fixation: Harvest cells and fix immediately in a mixture of 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1M sodium cacodylate buffer (pH 7.4) for a minimum of 1 hour at 4°C. Primary fixation is critical for preserving delicate early apoptotic structures.
- Washing: Rinse cells 3x with 0.1M cacodylate buffer.
- Post-fixation: Apply 1% osmium tetroxide in the same buffer for 1 hour at 4°C to stain lipid membranes and enhance contrast.
- Dehydration: Subject samples to a graded series of ethanol (50%, 70%, 90%, 100%) followed by a transition solvent like propylene oxide.
- Embedding: Infiltrate and embed samples in a resin such as Epon or Spurr's. Polymerize at 60°C for 48 hours.
Imaging and Analysis:
- Sectioning: Prepare ultrathin sections (60-90 nm) using an ultramicrotome and collect on copper grids. Counterstain with uranyl acetate and lead citrate.
- Blinded Evaluation: Acquire micrographs systematically at a standard primary magnification (e.g., 3000-8000x). The evaluator should be blinded to the experimental conditions to prevent bias.
- Scoring: Use a binary checklist (based on Table 1) to score each cell for the presence or absence of key apoptotic features. A cell is typically classified as apoptotic if it displays two or more definitive markers (e.g., chromatin margination and cell blebbing). Analyze a minimum of 100 cells per condition to ensure statistical power.
Molecular Pathways and Image Analysis Correlates
Understanding the molecular biology of apoptosis provides context for the morphological changes observed via TEM and guides the use of specific fluorescent probes and biochemical assays in correlative light and electron microscopy (CLEM) workflows.
Diagram 2: Apoptosis signaling pathways and effects (55 characters)
Correlative Imaging Protocols
Linking molecular events to ultrastructure is powerful. The optogenetic induction of apoptosis, as demonstrated with the OptoBAX system, is a prime example [52].
Protocol: Light-Activated Apoptosis Induction & Correlation (OptoBAX 2.0 System)
- Principle: A blue light-sensitive Cry2/CIB dimerization system recruits a BAX mutant (S184E) to the mitochondrial membrane, triggering MOMP.
- Transfection: Transfect cells (e.g., HEK293T, Neuro-2a) with constructs for
Cry2(1-531).L348F.mCh.BAX.S184EandTom20.CIB.GFP. - Induction & Live-Cell Imaging: Expose transfected cells to pulses of 470 nm blue light. Monitor in real-time using live-cell imaging for GFP/Mcherry localization, mitochondrial potential dyes (e.g., TMRM), and caspase activity sensors (e.g., FLICA).
- TEM Correlation: At defined timepoints post-induction (e.g., 30, 60, 120 mins), immediately fix the same cells for TEM processing as described in Section 2.1. This allows direct correlation of early molecular events (BAX translocation, caspase activation) with the ensuing ultrastructural changes.
The Scientist's Toolkit: Research Reagent Solutions
A curated set of reagents and tools is essential for executing the experiments described in this guide.
Table 2: Essential Reagents for Apoptosis Imaging and Analysis
| Reagent / Tool | Specific Example | Primary Function in Apoptosis Detection |
|---|---|---|
| Optogenetic Inducer | OptoBAX 2.0 (Cry2(1-531).L348F.mCh.BAX.S184E + Tom20.CIB.GFP) | Controlled, light-activated initiation of intrinsic apoptosis via BAX-mediated MOMP [52] |
| Caspase Activity Probe | FLICA (Fluorescent-Labeled Inhibitor of Caspases) | Fluorescently labels active caspase enzymes in live cells, providing a readout of key apoptotic protease activity |
| Membrane Integrity & PS Exposure | Annexin V-FITC / Propidium Iodide (PI) | Flow cytometry or fluorescence microscopy to detect phosphatidylserine externalization (early apoptosis) and loss of membrane integrity (late apoptosis/necrosis) [51] [14] |
| DNA Fragmentation Assay TUNEL (Terminal deoxynucleotidyl transferase dUTP Nick End Labeling) | In situ labeling of DNA strand breaks, a hallmark of late-stage apoptosis [24] | |
| Mitochondrial Dye | Tetramethylrhodamine, Methyl Ester (TMRM) | Assess mitochondrial membrane potential; depolarization occurs during intrinsic apoptosis |
| Image Analysis Software | CellProfiler, FIJI/ImageJ | Automated, high-throughput quantification of cellular morphology, fluorescence intensity, and object counting from microscopy images [53] [54] |
| Electron Microscopy Stains | Uranyl Acetate, Lead Citrate | Heavy metal stains used in TEM to generate contrast, highlighting membranes and nucleic acids in ultrathin sections [26] |
Standardized Reporting Checklist for Image Data
To ensure the reproducibility and clarity of published apoptosis image data, adherence to community-developed reporting standards is critical [54].
- Image Formatting (Minimal): Clearly indicate any cropping, rotation, or splicing of images. Use scale bars on all micrographs. Report the number of biological replicates (N) and the total cells (n) analyzed.
- Image Colors and Channels (Recommended): For fluorescence, provide channel annotations (e.g., "Caspase-3, red") and use accessible color palettes. For TEM, state the staining protocol used.
- Data Availability (Ideal): Make original, unprocessed image data (both fluorescence and TEM) available in a public repository. Share image analysis scripts and workflows (e.g., CellProfiler pipelines) to enable full reproducibility.
The systematic integration of standardized TEM analysis with molecular biochemistry and rigorous image quantification creates a powerful, multi-faceted toolkit for the definitive identification of early apoptosis. By adhering to the detailed protocols, checklists, and standardized reporting frameworks outlined in this guide, researchers can generate highly reliable, quantitative, and reproducible data on cell death mechanisms, thereby accelerating research in drug development, toxicology, and fundamental cell biology.
This case study details a practical investigation into the early detection of apoptotic human lens epithelial cells (LECs) using transmission electron microscopy (TEM). The research is situated within a broader thesis on the role of programmed cell death in the pathogenesis of senile cataracts, a leading cause of global blindness. The lens epithelium, a single layer of cells on the anterior lens surface, is essential for maintaining lens homeostasis and transparency; its degeneration via apoptosis is a critical early event in cataract formation [13] [55]. This study leverages the high-resolution capabilities of TEM to identify ultrastructural markers of early apoptosis in LECs obtained from patients undergoing cataract surgery, providing a methodological framework for similar research in ophthalmic cell biology and drug development.
Experimental Protocols and Methodologies
Patient Cohort and Sample Collection
An observational, cross-sectional study was conducted involving 21 Greek patients diagnosed with senile cataracts, all over the age of 60. Anterior lens capsules (aLCs), which include the monolayer of LECs, were carefully excised during routine phacoemulsification surgery via a continuous curvilinear capsulorrhexis technique. The samples, measuring 5-5.5 mm in diameter, were immediately fixed upon removal to preserve cellular integrity for subsequent ultrastructural analysis [13].
Sample Preparation for Transmission Electron Microscopy
Proper sample preparation is paramount for the accurate preservation of subcellular morphology. The following protocol was meticulously followed [13]:
- Primary Fixation: Samples were fixed in a neutral-buffered 3% glutaraldehyde solution for 90 minutes at room temperature. This cross-links proteins and stabilizes cellular structures.
- Post-fixation: Specimens were transferred to a 2% osmium tetroxide (OsO4) solution. Osmium tetroxide acts as a secondary fixative that stabilizes lipids and provides electron density to membranes.
- Dehydration: Fixed samples were dehydrated through a graded series of ethanol concentrations to remove all water from the tissue.
- Embedding: Dehydrated specimens were embedded in Epon 812 resin, which polymerizes to form a hard block suitable for thin-sectioning.
- Sectioning: Semi-thin sections (1-3 μm) were first cut and stained with 1% cyanetoluidine for initial examination under light microscopy. Subsequently, ultrathin sections (60-80 nm) were cut using an ultramicrotome.
- Staining: Ultrathin sections were double-stained with uranyl acetate and lead citrate to enhance contrast for TEM imaging.
- Imaging and Analysis: Sections were examined using a JEOL JEM-1011 transmission electron microscope operating at 80 kV. For each case, at least six different areas of the aLCs were examined, encompassing a minimum of 25 cells per sample to ensure a representative analysis [13].
Pre-embedding Immunogold Labeling Protocol
While not used in the primary apoptosis study, immunogold labeling is a powerful complementary technique for localizing specific proteins at the EM level. A standardized protocol for pre-embedding immunogold is as follows [56]:
- Fixation: Fix samples with 4% paraformaldehyde (PF) in phosphate-buffered saline (PBS) for 30 minutes. For some antibodies, including a low concentration of glutaraldehyde (0.05–0.2%) may improve structural preservation.
- Permeabilization and Blocking: Incubate fixed samples with a solution containing 5% normal goat serum (NGS) and 0.1% saponin in PBS to permeabilize membranes and block non-specific binding.
- Primary Antibody Incubation: Incubate with a primary antibody specific to the protein of interest.
- Secondary Antibody Incubation: Incubate with a Fab' fragment secondary antibody conjugated to 1.4 nm gold particles.
- Silver Enhancement: Treat samples with a silver enhancement kit to increase the size of the gold particles, making them visible under EM.
- Post-fixation and Processing: Post-fix with osmium tetroxide, followed by standard dehydration, embedding, and sectioning procedures as outlined above [56].
Results and Data Analysis
Ultrastructural Identification of Apoptotic Lens Epithelial Cells
The core finding of this study was the identification of apoptotic LECs based on definitive morphological criteria established by TEM. Apoptotic cells were detected in 9 out of the 21 patients (42.9%) [13]. The key ultrastructural features observed are summarized in the table below and form the basis for identifying early apoptosis.
Table 1: Key Ultrastructural Features of Apoptotic Human Lens Epithelial Cells Identified by TEM
| Cellular Compartment | Morphological Hallmarks of Apoptosis | Frequency / Notes |
|---|---|---|
| Nucleus | Chromatin condensation and margination, nuclear membrane irregularity, reduction of nuclear volume (pyknosis), nuclear degradation (karyorrhexis) | Universally observed in apoptotic LECs; nuclei often appeared uniformly dense or empty [13] |
| Cytoplasm | Increased electron density, presence of numerous cytoplasmic vacuoles, reduction in mitochondria, expansion of rough endoplasmic reticulum | Cytoplasm was sometimes dense but most often contained vacuoles [13] |
| Plasma Membrane | Membrane blebbing, budding, presence of pinopode-like projections, loose or no connections with neighboring cells and the basement membrane | Frequently observed; apoptotic cells appeared smaller and isolated [13] |
| Apoptotic Bodies | Membrane-bound vesicles containing tightly packed organelles and/or nuclear fragments | Detected in a few cases; phagocytosed by neighboring LECs [13] |
The following diagram illustrates the key morphological features of an apoptotic Lens Epithelial Cell and its subsequent phagocytic clearance, creating a visual summary of the process described in the results.
Statistical Correlation with Risk Factors
The study statistically analyzed the presence of apoptotic LECs against several patient and clinical variables. The results demonstrated that none of the examined risk factors—including age, gender, biomicroscopic cataract type, and the coexistence of exfoliation syndrome (XFS), diabetes mellitus, or glaucoma—showed a statistically significant connection with the incidence of apoptosis in this cohort [13].
Table 2: Statistical Analysis of Apoptosis Correlation with Patient Factors
| Variable | Group Comparison | p-value |
|---|---|---|
| Exfoliation Syndrome (XFS) | Apoptosis in 6/11 XFS patients vs. 3/10 non-XFS patients | 0.575 |
| Gender | Apoptosis in 4/7 males vs. 5/14 females | 0.642 |
| Cataract Type | Distribution across cortical, nuclear, and posterior subcapsular types | 0.368 |
| Age | Mean age 70.77 (with apoptosis) vs. 73.41 (without apoptosis) | 0.468 |
| Diabetes Mellitus | Apoptosis in 3/6 patients with DM vs. 3/10 without DM | 0.523 |
A pivotal observation was the role of neighboring cells in the apoptotic process. Neighboring LECs were found to phagocytose the apoptotic bodies, effectively functioning as macrophages to clear the dying cells from the tissue. This process prevents the release of cellular contents and secondary necrosis, which is crucial for maintaining tissue integrity and a non-inflammatory state [13].
The Scientist's Toolkit: Essential Research Reagents and Materials
Successful TEM research on LECs requires a specific set of reagents and materials. The following table details key items used in the featured experiments and their critical functions.
Table 3: Key Research Reagent Solutions for TEM-based Apoptosis Detection in LECs
| Reagent / Material | Function in Protocol |
|---|---|
| Glutaraldehyde (3%) | Primary fixative; cross-links proteins to preserve ultrastructure. |
| Osmium Tetroxide (OsO4) | Post-fixative; stabilizes lipids and adds electron density to membranes. |
| Epon 812 Resin | Embedding medium; infiltrates tissue and hardens for thin-sectioning. |
| Uranyl Acetate & Lead Citrate | Heavy metal stains; bind to cellular components to enhance image contrast. |
| Primary Antibodies (e.g., Anti-CRYAB) | For immunogold; specifically binds to target proteins (e.g., crystallins) [56] [57]. |
| Nanogold-Fab' Secondary Antibody | For immunogold; Fab' fragment conjugated to 1.4 nm gold particles binds primary antibody [56]. |
| Silver Enhancement Kit | For immunogold; deposits metallic silver onto gold particles, amplifying signal size [56]. |
| Paraformaldehyde (4%) | Fixative for immunogold labeling; provides good antigen preservation [56]. |
| Saponin (0.1%) | Detergent for immunogold; permeabilizes cell membranes to allow antibody entry [56]. |
Discussion and Workflow Integration
This case study demonstrates that TEM remains the gold standard for the unequivocal identification of early apoptotic cells based on morphological criteria, a claim supported by its ability to reveal specific features like chromatin margination and membrane blebbing that are hallmarks of this cell death pathway [13]. The finding that 42.9% of senile cataract patients exhibited apoptotic LECs underscores the potential significance of this process in cataractogenesis, even in the absence of a statistical link to the common risk factors examined.
The experimental workflow for detecting apoptosis in LECs, from sample acquisition to image interpretation, can be integrated into a single, coherent pipeline. This workflow also highlights how complementary techniques can be incorporated to enrich the analysis, as illustrated in the following diagram.
The phagocytosis of apoptotic bodies by neighboring LECs, as observed in this study, is a critical mechanism for the non-inflammatory clearance of dead cells. A recent groundbreaking study has identified a related mechanism involving the "FOotprint Of Death" (FOOD), where retracting apoptotic cells leave behind actin-rich, membrane-bound footprints on the substrate that vesicularize into large extracellular vesicles (F-ApoEVs). These F-ApoEVs expose "eat-me" signals like phosphatidylserine, effectively marking the site of cell death for phagocytes and facilitating efferocytosis [17]. This discovery provides a novel framework for understanding how apoptotic LECs might communicate with their environment to ensure their own tidy removal.
For future research, the integration of advanced techniques holds great promise. Correlative light and electron microscopy (CLEM) can bridge the gap between dynamic live-cell imaging and high-resolution ultrastructure [58]. Furthermore, computational methods like the deep learning-based apoptosis detection system (ADeS) have been developed to automatically detect apoptosis in live-cell imaging with high accuracy, offering a powerful tool for screening and quantification [59]. These methodologies, combined with the foundational TEM protocols outlined herein, will continue to advance our understanding of cell death mechanisms in the lens and their implications for drug development and therapeutic interventions for cataract and other diseases.
Optimizing Results: Troubleshooting Common Artifacts and Technical Challenges in TEM
Transmission electron microscopy (TEM) remains the gold standard for the ultrastructural identification of early apoptotic cells, revealing characteristic morphology such as chromatin condensation, nuclear membrane budding, and cytoplasmic vacuolization. However, the intricate processes of chemical fixation, dehydration, and staining during sample preparation can introduce artifacts that closely mimic or obscure these key apoptotic features. Misinterpretation can lead to false positives or negatives, potentially compromising experimental results in cell biology research and drug development. This guide provides an in-depth analysis of common TEM artifacts relevant to apoptosis studies, supported by quantitative data and detailed protocols to enhance the accuracy of your morphological assessments.
Quantitative Analysis of Artifacts and Apoptotic Features
A systematic approach to identification requires an understanding of the prevalence and distinguishing features of both artifacts and true apoptosis. The following tables summarize key quantitative data and morphological characteristics to aid in this differentiation.
Table 1: Prevalence and Impact of Common TEM Artifacts in Apoptosis Studies
| Artifact Type | Primary Cause | Mimicked Apoptotic Feature | Estimated Frequency in Poorly Prepared Samples | Key Distinguishing Characteristic |
|---|---|---|---|---|
| Cytoplasmic Shrinkage | Osmolarity imbalance in fixative | Cell shrinkage, a classic early apoptotic sign | 40-60% | Formation of clear, empty spaces between plasma membrane and organelles, unlike apoptotic condensation. |
| Nuclear Scalloping | Incomplete perfusion or slow fixation | Nuclear membrane budding and blebbing | 15-30% | Irregular, coarse indentation of the entire nuclear envelope, unlike the discrete, rounded apoptotic blebs. |
| Mitochondrial Swelling | Hypoxia or chemical toxicity before fixation | Early apoptotic mitochondrial changes | 10-25% | Extreme matrix expansion and rupture of cristae, unlike the condensed morphology in early apoptosis. |
| Chromatin Clumping | Poor buffer pH or delayed fixation | Chromatin condensation and margination | 20-35% | Coarse, irregular clumping distributed throughout the nucleus, unlike the smooth, peripheral condensation in apoptosis. |
Table 2: Distinguishing True Early Apoptosis from Common Artifacts
| Cellular Feature | True Early Apoptosis | Common Artifact | Diagnostic Cue for TEM |
|---|---|---|---|
| Cell Membrane | Preservation of membrane integrity with budding into apoptotic bodies. | Detachment and tearing, creating empty peri-cellular spaces. | Look for intact, vesiculating membranes versus torn or separated ones. |
| Chromatin | Smooth, uniform condensation along the inner nuclear membrane. | Irregular, coarse clumping throughout the nucleoplasm. | Assess pattern: peripheral and uniform is key for true apoptosis. |
| Cytoplasm | Condensation with organelle integrity largely preserved. | Gross vacuolization or organelle-free swelling. | Organelles remain functional-looking; density increases. |
| Mitochondria | Condensed configuration with dark matrix. | Swollen, translucent matrix with broken cristae. | Matrix density is a critical differentiator. |
Experimental Protocols for Optimal Sample Preparation
Accurate visualization of apoptosis requires meticulous sample preparation to minimize artifacts. The following protocols are optimized for preserving true cellular morphology.
Protocol for Buffered Glutaraldehyde Primary Fixation
This protocol ensures rapid and uniform fixation, crucial for preserving the dynamic morphology of early apoptosis [60].
Preparation of Fixative Solution:
- Prepare a 2.5% glutaraldehyde solution in 0.1M sodium cacodylate buffer (pH 7.4).
- Verify the osmolarity of the fixative is appropriate for your cell type (typically ~300 mOsm). Adjust with sucrose or sodium chloride if necessary.
- Pre-cool the fixative to 4°C.
Fixation Procedure:
- For Cell Cultures: Gently add pre-cooled fixative directly to the culture medium to achieve a 1:1 mixture. After 2 minutes, replace the mixture with fresh, pure fixative. This gradual introduction minimizes osmotic shock.
- For Tissues: Perform vascular perfusion for optimal results. If immersion fixation is necessary, dissect tissue into <1 mm³ pieces and immerse immediately in cold fixative.
Fixation Duration: Fix samples for a minimum of 2 hours at 4°C [60].
Protocol for Correlative Light and Electron Microscopy (CLEM)
A 3D-CLEM workflow, as established for studying nanoparticle uptake, can be adapted to unambiguously identify apoptotic cells by correlating dynamic fluorescent markers (e.g., Annexin V, caspase sensors) with ultrastructural detail [60].
Live-Cell Imaging and Marking:
- Culture cells on a gridded, glass-bottom dish suitable for both light and electron microscopy.
- Induce apoptosis and stain with a fluorescent marker (e.g., FLICA for caspase activity).
- Using confocal fluorescence microscopy, identify and record the coordinates of fluorescent-positive cells of interest within the grid.
Correlative Processing for TEM:
- Fix the entire sample using the protocol in 2.1, but with the addition of a low-concentration fluorescent dye (e.g, 0.1% Eosin Y) in the buffer to aid in relocating cells post-embedding.
- Process the sample through standard dehydration and resin embedding (e.g., Epon) steps directly on the dish.
- After polymerization, use the grid coordinates and residual fluorescence to locate the exact cells previously identified by light microscopy.
- Trim the resin block around the cell of interest and prepare ultrathin sections for TEM imaging.
Image Correlation and Analysis:
- Correlate the fluorescence image with the TEM micrograph. This confirms that the ultrastructure being analyzed definitively comes from a cell that was in the early stages of apoptosis, eliminating ambiguity from artifacts [60].
Diagram 1: CLEM workflow for definitive apoptosis identification.
The Scientist's Toolkit: Essential Reagents and Materials
Table 3: Research Reagent Solutions for Apoptosis TEM Studies
| Reagent / Material | Function / Application | Critical Parameter / Rationale |
|---|---|---|
| Glutaraldehyde (EM Grade) | Primary fixative for cross-linking proteins, stabilizing structure. | High purity (≥25%, EM grade) is essential to prevent polymerized contaminants that cause background noise. |
| Sodium Cacodylate Buffer | Buffering system for primary fixative. | Maintains stable physiological pH (7.2-7.4) during fixation; superior to phosphate buffers for preventing precipitate formation. |
| Osmium Tetroxide | Secondary fixative that stabilizes lipids and imparts electron density. | Critical for preserving membrane structures (nuclear envelope, mitochondria). Must be handled in a fume hood. |
| Uranyl Acetate | Heavy metal stain for en bloc or section staining. | Enhances contrast of nucleic acids and membranes. Lead citrate is used subsequently for general contrast. |
| Epon / Epoxy Resin | Embedding medium for ultrathin sectioning. | Provides mechanical stability for cutting ~70 nm sections. Formulation must be meticulously followed for proper polymerization. |
| Gridded Coverslip Dish | Substrate for CLEM workflows. | Allows for precise relocation of the same cell from light microscopy to TEM, enabling definitive correlation [60]. |
Logical Framework for Differentiating Apoptosis from Artifacts
A systematic decision-making process is required when analyzing cellular ultrastructure. The following diagram outlines the key questions to ask when evaluating ambiguous morphology.
Diagram 2: Diagnostic logic for apoptosis versus artifact.
The accurate identification and quantification of early apoptosis are critical for research in cell biology, oncology, and drug development. The transient nature of early apoptotic events and the low frequency at which they occur in cellular populations present significant methodological challenges [3]. During early apoptosis, cells undergo a series of characteristic morphological and biochemical changes, including cell shrinkage, phosphatidylserine (PS) externalization, and chromatin condensation, while maintaining membrane integrity [3]. However, this phase is remarkably brief, often making it difficult to capture with standard endpoint assays. Furthermore, in many experimental systems, only a small percentage of the cell population may be undergoing apoptosis at any given time, necessitating highly sensitive detection methods capable of identifying these rare events against a background of viable cells [37].
These challenges are particularly acute in transmission electron microscopy (TEM) research, where the ultrastructural assessment of apoptosis provides the definitive morphological criteria for its identification [3]. This technical guide examines the core challenges in quantifying early apoptosis and provides detailed methodologies and reagent solutions to enhance detection accuracy within the context of TEM-based research.
Core Challenges in Detection and Quantification
The Transient Nature of Early Apoptotic Events
The progression from early to late apoptosis occurs rapidly, creating a narrow window for detection. Early features like PS externalization and cell shrinkage may be visible for only a few hours before the cell advances to later stages involving DNA fragmentation and apoptotic body formation [3]. This temporal dynamic means that single time-point measurements often fail to capture the full extent of apoptosis in a population. As noted in assessments of melanoma stem cells, apoptotic responses can exhibit distinct temporal dynamics between different cell types, with some populations showing delayed but sustained increases in caspase activation [61]. This underscores the need for time-course experiments rather than single endpoint measurements to accurately quantify apoptotic induction.
Low Frequency of Occurrence in Cellular Populations
In many experimental models, particularly those involving low-level stress or chemotherapeutic agents, the proportion of cells simultaneously undergoing early apoptosis may be very small. This low frequency compounds the challenges of detection, as analytical techniques must be capable of identifying these rare events with high specificity and sensitivity [37]. Flow cytometry and microscopy-based methods face statistical limitations when trying to accurately quantify small subpopulations, potentially leading to both false negatives and unreliable quantitative data.
Technical Limitations of Classic Detection Methods
Classic apoptosis detection methods, including DNA-binding dyes like DAPI and PI, frequently lack the specificity required to definitively distinguish early apoptotic cells from viable or necrotic cells [37]. As highlighted in recent methodological reviews, "DAPI and PI cannot distinguish between early apoptotic and necrotic cells leading to potential false-positive results when used to detect apoptosis" [37]. Furthermore, Annexin V staining, while more specific for PS externalization, relies on expensive recombinant proteins and may introduce artifacts [37]. The lack of real-time monitoring capabilities in most classic methods further complicates the capture of transient apoptotic events as they occur.
Table 1: Challenges and Consequences in Early Apoptosis Quantification
| Challenge | Impact on Quantification | Technical Consequences |
|---|---|---|
| Transient Nature | Narrow detection window for early biomarkers | Single time-point measurements yield false negatives; requires kinetic assays |
| Low Frequency | Rare events difficult to detect statistically | Reduced signal-to-noise ratio; requires high-sensitivity methods |
| Morphological Continuum | Continuum between apoptosis and necrosis | Difficulty in definitive classification; requires multi-parameter assessment |
Current Detection Strategies and Their Applications
Morphological Assessment via Transmission Electron Microscopy
TEM provides the gold standard for definitive identification of apoptotic cells based on ultrastructural morphology, making it particularly valuable for validating other detection methods [3]. The key early apoptotic features identifiable by TEM include cell shrinkage, chromatin condensation into crescent-shaped masses at the nuclear periphery, and membrane blebbing while maintaining membrane integrity [3]. As Leist and Jaättela describe, the specific morphology of condensed chromatin can even provide information about the biochemical pathway involved, with "caspase-dependent apoptosis mostly induc[ing] strong chromatin compaction in crescent shaped masses at the nuclear periphery" [3].
The primary advantage of TEM in addressing the quantification challenges lies in its unparalleled resolution for identifying the cardinal features of early apoptosis. However, its limitations for quantification include the small sample area that can be analyzed and the labor-intensive nature of sample preparation and image analysis [3]. Thus, while TEM provides definitive qualitative assessment, it is often combined with other higher-throughput methods for quantitative analysis.
Luminescence and Fluorescence-Based Advanced Methods
Innovative luminescence-based methods have emerged to address the limitations of classic techniques, offering increased sensitivity, pathway specificity, and negligible cytotoxicity [37]. These approaches are particularly valuable for detecting low-frequency events and for real-time monitoring of apoptotic progression. Split luciferase complementation assays, for instance, can detect apoptosome formation in real-time, allowing researchers to track the dynamic process of caspase activation rather than relying on single endpoint measurements [37].
Fluorescence-based techniques using carbon dots-annexin V probes and fluorometric assays for cytochrome c release provide enhanced sensitivity for detecting early apoptotic markers [37]. These methods are particularly useful for addressing the low-frequency challenge, as they can identify rare events within large cell populations. The development of caspase-specific fluorescent inhibitors and aggregation-induced emission luminogens further allows for precise tracking of enzyme activation central to the apoptotic process [37].
Table 2: Biomarkers and Advanced Methods for Early Apoptosis Detection
| Apoptosis Stage | Key Biomarkers | Advanced Detection Methods | Advantages for Quantification Challenges |
|---|---|---|---|
| Early | Phosphatidylserine externalization | Carbon dots-annexin V probes & fluorescence spectroscopy [37] | High sensitivity for low-frequency events |
| Mitochondrial membrane depolarization | Membrane-permeable fluorescent dyes & fluorescence microscopy [37] | Early detection before caspase activation | |
| Caspase activation | Split luciferase complementation assays [37] | Real-time monitoring of transient events | |
| Chromatin condensation | High-resolution confocal imaging – single molecule localization microscopy [37] | High-resolution spatial analysis |
Flow Cytometry and Multi-Parameter Approaches
Flow cytometry represents a powerful tool for addressing both the transient nature and low frequency of early apoptosis through its capacity for multi-parameter analysis of individual cells within large populations [37]. By simultaneously measuring multiple parameters such as PS externalization (via Annexin V binding), membrane integrity (via dye exclusion), caspase activation, and mitochondrial membrane potential, researchers can more confidently identify and quantify the small proportion of cells in early apoptosis [37].
Recent advancements in imaging flow cytometry combined with convolutional autoencoders have further enhanced the ability to detect subtle morphological changes characteristic of early apoptosis, bridging the gap between traditional flow cytometry and morphological assessment [37]. This approach is particularly valuable for capturing the transient nature of early apoptosis, as it allows for the analysis of rapid kinetic processes in cell populations.
Experimental Protocols for Enhanced Quantification
Integrated Workflow for TEM and Flow Cytometry
To overcome the individual limitations of different detection methods, an integrated protocol combining flow cytometry with TEM validation provides a robust approach for accurate quantification of early apoptosis:
Sample Preparation: Treat cells with apoptotic inducers (e.g., staurosporine, etoposide) across multiple time points to capture kinetic progression [62]. Include appropriate controls (untreated cells) and consider glucose deprivation conditions to modulate ATP-dependent apoptosis [62].
Multi-Parameter Flow Cytometry:
- Harvest cells by gentle trypsinization or use floating cells in the supernatant.
- Stain with Annexin V-FITC in binding buffer for 15 minutes in the dark [37].
- Add propidium iodide (PI) immediately before analysis to distinguish early apoptotic (Annexin V+/PI-) from late apoptotic/necrotic cells (Annexin V+/PI+) [37].
- For enhanced early detection, include fluorescent caspase inhibitors (e.g., FAM-DEVD-FMK) or mitochondrial membrane potential dyes (e.g., JC-1) [37].
- Analyze using a flow cytometer with appropriate filters, collecting data for at least 10,000 events per sample.
TEM Sample Preparation:
- Pellet cells and fix with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for 2 hours at 4°C [3].
- Post-fix with 1% osmium tetroxide for 1 hour, then dehydrate through a graded ethanol series.
- Embed in epoxy resin and polymerize at 60°C for 48 hours.
- Prepare ultrathin sections (70-90 nm) using an ultramicrotome and counterstain with uranyl acetate and lead citrate [3].
- Examine using TEM at 80 kV, focusing on ultrastructural features of early apoptosis.
Data Correlation: Correlate flow cytometry data with TEM findings to validate the identification of early apoptotic cells and establish quantitative correlations between biochemical and morphological markers.
Real-Time Kinetic Apoptosis Assay Using Bioluminescence
For monitoring the transient nature of early apoptosis in real-time, a bioluminescence-based ATP monitoring assay can be employed, as apoptosis requires energy and exhibits increased cytosolic ATP levels:
Cell Preparation: Transfect cells with the firefly luciferase gene using an appropriate vector system to enable cytosolic ATP monitoring [62].
Bioluminescence Recording:
- Seed luciferase-transfected cells in specialized culture dishes compatible with bioluminescence recording.
- Add luciferin substrate (0.5 mM) to the culture medium 1 hour before measurements.
- Place cells in a dark chamber at 37°C with 5% CO2 and record basal bioluminescence for 30 minutes.
- Add apoptotic inducer (e.g., staurosporine, TNFα) without interrupting recording.
- Continue bioluminescence monitoring for up to 6 hours, as "the cytosolic ATP level remained at a higher level than in the control for up to 6 h during which activation of caspase-3 and internucleosomal DNA fragmentation took place" [62].
Data Interpretation: Normalize bioluminescence signals to baseline and monitor for sustained increases in cytosolic ATP, which indicates progression through apoptosis.
High-Content Analysis for Low-Frequency Detection
For detecting low-frequency early apoptosis, a high-content screening approach using automated microscopy provides both statistical power and morphological validation:
Cell Preparation and Staining:
- Seed cells in 96-well or 384-well imaging plates and treat with experimental conditions.
- At appropriate time points, stain with Hoechst 33342 (nuclear staining), Annexin V-Alexa Fluor 488 (PS externalization), and MitoTracker Deep Red (mitochondrial mass) [37].
- Include a caspase-3/7 activation fluorogenic substrate for additional specificity.
Automated Imaging and Analysis:
- Image plates using a high-content imaging system with environmental control to maintain cell viability during extended imaging.
- Acquire images at multiple time points (every 2-4 hours) to capture kinetic progression.
- Use automated image analysis software to identify early apoptotic cells based on multiple parameters: condensed/fragmented nuclei, positive Annexin V staining, maintained membrane integrity, and mitochondrial morphology.
Statistical Analysis: Apply statistical methods appropriate for rare event detection, with confidence intervals calculated based on the total number of cells analyzed per condition.
Signaling Pathways in Early Apoptosis
The molecular pathways of apoptosis involve sophisticated regulatory mechanisms that present both challenges and opportunities for detection. The following diagram illustrates the key pathways and their interconnections:
Diagram 1: Apoptosis Signaling Pathways. The diagram illustrates the major pathways of apoptosis induction, highlighting key detection points. The recently identified VDAC1-Bcl-xL regulatory switch is shown as a critical control mechanism in the mitochondrial pathway [63].
The diagram above illustrates the complex interplay between the extrinsic (death receptor) and intrinsic (mitochondrial) apoptotic pathways. Recent research has identified novel regulatory switches in these pathways, such as the VDAC1 protein which "unfolds part of its structure, connects it to Bcl-xL, and thus deactivates the inhibitor" under cellular stress conditions [63]. Understanding these pathways is essential for developing targeted detection strategies, as different stimuli activate apoptosis through distinct mechanisms with varying kinetics.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Research Reagent Solutions for Apoptosis Detection
| Reagent/Category | Specific Examples | Function in Apoptosis Detection | Considerations for Use |
|---|---|---|---|
| PS Binding Agents | Annexin V-FITC, Carbon dots-annexin V probes [37] | Detects phosphatidylserine externalization on outer membrane leaflet | Requires calcium-containing buffer; use with viability dye to exclude late apoptotic/necrotic cells |
| Caspase Substrates | FAM-DEVD-FMK, Luminescent caspase substrates [37] | Detects activation of executioner caspases (3/7) | Distinguish between early (activation) and late (decline) apoptosis; use cell-permeable inhibitors for live-cell imaging |
| Mitochondrial Dyes | JC-1, MitoTracker Red CMXRos, MitoSOX Red [37] | Measures mitochondrial membrane potential and ROS production | Critical for detecting early intrinsic pathway activation; optimize loading conditions for each cell type |
| Nuclear Stains | Hoechst 33342, DAPI, FxCycle PI/RNase [3] | Identifies chromatin condensation and nuclear fragmentation | Hoechst 33342 is cell-permeable for live cells; DAPI requires permeabilization |
| ATP Monitoring | Luciferase-transfected cells + luciferin [62] | Monitors cytosolic ATP levels which increase during apoptosis | Requires genetic modification but enables real-time kinetic measurements |
| Apoptosis Inducers | Staurosporine, Etoposide, TNFα with CHX [62] | Positive controls for inducing apoptosis through various pathways | Optimize concentration and time course for each cell type; include in validation experiments |
The accurate quantification of early apoptosis remains challenging due to the transient nature of the process and the frequently low frequency of apoptotic cells in experimental systems. Addressing these challenges requires integrated methodological approaches that combine the ultrastructural validation provided by TEM with sensitive biochemical and molecular detection techniques. The development of real-time monitoring systems and advanced luminescence-based methods has significantly improved our ability to capture the dynamic progression of apoptosis, while multi-parameter flow cytometry and high-content analysis enhance the statistical power for detecting low-frequency events.
Future directions in apoptosis quantification will likely focus on further refining kinetic assessment methods and developing more specific probes for early apoptotic markers. The continued elucidation of novel regulatory mechanisms, such as the recently identified VDAC1-Bcl-xL switch [63], will provide additional molecular targets for precise detection and quantification. By implementing the comprehensive strategies and methodologies outlined in this technical guide, researchers can significantly improve the accuracy and reliability of early apoptosis quantification in both basic research and drug development contexts.
Within the context of a broader thesis on transmission electron microscopy (TEM) identification of early apoptosis, this guide provides a detailed technical framework for researchers aiming to accurately differentiate apoptosis from autophagy and other forms of regulated cell death (RCD). The intricate crosstalk and overlapping molecular regulators among these pathways make specific distinction a critical challenge in cell death research [64]. This document outlines definitive strategies based on morphological, biochemical, and functional criteria, providing standardized protocols and analytical tools to ensure specificity in experimental findings, thereby enhancing the reliability of research in drug development and mechanistic studies.
Morphological Hallmarks: The First Line of Distinction
Morphological assessment, particularly using transmission electron microscopy (TEM), provides the most definitive evidence for distinguishing different forms of cell death by revealing unique and unambiguous ultrastructural features [65] [66].
Table 1: Morphological Characteristics of Apoptosis, Autophagy, and Other RCD Forms
| Cell Death Type | Nuclear Morphology | Cytoplasmic and Organellar Features | Plasma Membrane | TEM Identification Key |
|---|---|---|---|---|
| Apoptosis | Chromatin condensation (pyknosis), nuclear fragmentation (karyorrhexis), formation of apoptotic bodies [65] [66] | Cell shrinkage, concentrated cytoplasm, organelles often remain intact initially [65] | Membrane blebbing, formation of apoptotic bodies with intact membrane; no inflammatory response [66] | Early stage: Chromatin margination, cell shrinkage. Late stage: Apoptotic bodies containing nuclear fragments and organelles [65]. |
| Autophagy | Lack of chromatin condensation; nucleus may appear normal until late stages [67] [68] | Massive vacuolization, presence of double-membraned autophagosomes engulfing cytoplasmic content [67] [68] | Intact until late stages [64] | Identification of phagophores (cup-shaped double membranes), autophagosomes (double-membraned vesicles containing cytoplasm/organelles), and autolysosomes (single-membraned, electron-dense vesicles) [68]. |
| Necroptosis | Nuclear dehydration (pyknosis) but retains an integral nucleus [67] [66] | Cell and organelle swelling (oncosis), translucent cytoplasm, dilation of Golgi and ER [66] | Rupture and loss of integrity, release of pro-inflammatory cellular contents [67] [66] | Swollen organelles and ruptured plasma membrane without the formation of apoptotic bodies or autophagic vacuoles. |
| Pyroptosis | Nuclear condensation [66] | Cytoplasmic swelling [66] | Pore formation, swelling, and rupture with release of pro-inflammatory intracellular contents [67] [66] | Cells are enlarged with large plasma membrane pores before lysis. |
Experimental Protocol: TEM for Morphological Identification
Method: Transmission Electron Microscopy for Ultrastructural Analysis of Cell Death [65] [68]
- Sample Fixation: Fix cell pellets or tissue samples (1 mm³) in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for at least 2 hours at 4°C.
- Post-fixation: Wash samples with buffer and post-fix with 1% osmium tetroxide for 1-2 hours.
- Dehydration: Dehydrate samples through a graded series of ethanol (50%, 70%, 90%, 100%).
- Embedding: Infiltrate and embed samples in epoxy resin (e.g., Epon/Araldite) and polymerize at 60°C for 48 hours.
- Sectioning: Use an ultramicrotome to cut ultrathin sections (60-90 nm) and collect them on copper grids.
- Staining: Stain grids with uranyl acetate (for contrast) and lead citrate (to reduce electron scattering) [65].
- Imaging & Analysis: Observe under TEM at 80 kV. Systematically scan grids and capture micrographs. Identify cell death modalities by referencing Table 1, focusing on membrane structures and nuclear morphology.
Biochemical and Molecular Assays: Targeting Signature Biomarkers
Biochemical assays provide a quantitative and often stage-specific measure of cell death activity. Combining multiple assays is crucial for conclusive identification.
Table 2: Key Biochemical Biomarkers and Detection Assays
| Cell Death Type | Key Biomarkers | Detection Assays | Experimental Notes |
|---|---|---|---|
| Apoptosis | - Phosphatidylserine (PS) externalization [19]- Caspase-3/7, -8, -9 activation [66]- DNA fragmentation (180-200 bp ladder) [65] [66] | - Annexin V-FITC/PI flow cytometry (PS exposure) [19]- Western Blot for cleaved caspases, PARP [66]- TUNEL assay (DNA breaks) [65] [67]- DNA gel electrophoresis (DNA ladder) [65] | Annexin V+/PI- indicates early apoptosis; Annexin V+/PI+ indicates late apoptosis/secondary necrosis [19]. TUNEL can give false positives; use with controls [65]. |
| Autophagy | - LC3-I to LC3-II lipidation [68]- Increased levels of Beclin-1, ATG5 [68]- Degradation of p62/SQSTM1 [67] | - Western Blot for LC3-II/LC3-I ratio, p62 [68]- Immunofluorescence using GFP-LC3 or mRFP-GFP-LC3 reporters (punta formation) [69] [67] | LC3-II:LC3-I ratio and p62 degradation indicate autophagic flux. GFP-LC3 punta represent autophagosomes; lysosomal quenching of GFP in mRFP-GFP-LC3 indicates autolysosome formation. |
| Necroptosis | - Phosphorylation of RIPK1, RIPK3, and MLKL [67] [66] | - Western Blot for p-RIPK3, p-MLKL [67]- Immunoprecipitation of RIP1/RIP3 complex (necrosome) [67] | Phospho-MLKL oligomers form pores in the plasma membrane. Inhibition by Nec-1 (RIPK1 inhibitor) confirms necroptosis. |
| Pyroptosis | - Cleavage of Gasdermin D (GSDMD)- Activation of Caspase-1 (not Caspase-3) [66]- Release of IL-1β and IL-18 [67] | - Western Blot for cleaved GSDMD-NT fragment, cleaved Caspase-1 [67]- ELISA for extracellular IL-1β/IL-18 [67] | Dependent on inflammatory caspases (1/4/5 in humans, 11 in mice). Distinct from apoptotic caspase activation. |
Experimental Protocol: Annexin V/Propidium Iodide Assay by Flow Cytometry
Method: Flow Cytometric Analysis of Phosphatidylserine Externalization and Membrane Integrity [19]
- Cell Preparation: Harvest cells (e.g., by gentle trypsinization or collecting suspension cells) and wash twice with cold PBS.
- Staining: Resuspend 1 x 10⁵ to 1 x 10⁶ cells in 100 µL of 1X Annexin V Binding Buffer.
- Add 5 µL of FITC-conjugated Annexin V and 5 µL of Propidium Iodide (PI) staining solution (or appropriate volume as per manufacturer's instructions).
- Gently vortex the cells and incubate for 15 minutes at room temperature (25°C) in the dark.
- Analysis: Within 1 hour, add 400 µL of 1X Annexin V Binding Buffer to each tube and analyze by flow cytometry.
- Viable cells: Annexin V-/PI-
- Early Apoptotic cells: Annexin V+/PI-
- Late Apoptotic/Necrotic cells: Annexin V+/PI+
Functional and Pathway Analysis: Interrogating Molecular Mechanisms
Understanding the core signaling pathways and their crosstalk is essential for interpreting complex cell death scenarios. Cytosolic calcium (Ca²⁺) acts as a key rheostat, fine-tuning the decision between autophagy and apoptosis in response to stress levels [69].
Diagram: Calcium-Mediated Cell Fate Decision. Low-to-moderate stress promotes survival via Ca²⁺/CaMKKβ/AMPK-driven autophagy. High stress shifts the response to apoptosis via calpain activation and mitochondrial dysfunction. Caspases can inhibit autophagy by cleaving Beclin-1, reinforcing the death decision [69] [64].
Key Crosstalk Mechanisms and Molecular Switches
- Bcl-2 Family Proteins: The anti-apoptotic protein Bcl-2 inhibits apoptosis by maintaining mitochondrial membrane integrity. It also binds and inhibits Beclin-1, thereby suppressing autophagy. Stress-induced phosphorylation of Bcl-2 can release Beclin-1, simultaneously inducing autophagy and reducing the threshold for apoptosis [69] [64].
- p53 Dynamics: Nuclear p53 can promote apoptosis by transcribing pro-apoptotic genes like PUMA and Bax. Cytoplasmic p53, however, exerts a tonic inhibition on autophagy. The localization and status of p53 thus serve as a critical switch between the two pathways [69].
- Caspase-Mediated Inactivation of Autophagy: Executioner caspases (e.g., Caspase-3) cleave key autophagy proteins like Beclin-1, generating C-terminal fragments that further amplify apoptosis. This represents a point of no return, where apoptosis actively shuts down the pro-survival autophagy pathway [69] [64].
The Researcher's Toolkit: Essential Reagents and Materials
Table 3: Research Reagent Solutions for Distinguishing RCD
| Reagent / Assay Kit | Specific Target | Primary Function in Distinction | Key Considerations |
|---|---|---|---|
| Annexin V-FITC/PI Kit [19] | PS externalization (Apoptosis) | Labels early apoptotic cells (Annexin V+/PI-) | Requires live cells; PI stains necrotic/late apoptotic cells with compromised membranes. Standard for flow cytometry. |
| Z-VAD-FMK (Pan-Caspase Inhibitor) | Active caspases (Apoptosis) | Pharmacological inhibitor to confirm caspase-dependent apoptosis. | Failure of Z-VAD to inhibit cell death suggests a non-apoptotic, caspase-independent pathway (e.g., necroptosis). |
| Chloroquine / Bafilomycin A1 | Lysosomal function / Autophagic flux | Inhibits autophagosome-lysosome fusion, causing LC3-II accumulation. | Used to measure autophagic flux (difference in LC3-II with/without inhibitor) rather than just snapshot LC3 levels. |
| Necrostatin-1 (Nec-1) | RIPK1 (Necroptosis) | Specific inhibitor of receptor-interacting protein kinase 1 (RIPK1). | Confirms necroptosis when cell death is blocked by Nec-1 but not by Z-VAD. |
| Anti-LC3B Antibody | LC3 protein (Autophagy) | Detects LC3-I (cytosolic) and LC3-II (lipidated, autophagosome-bound) via WB or IF. | A increase in LC3-II:LC3-I ratio and appearance of puncta in IF indicate autophagy induction. |
| Anti-Cleaved Caspase-3 Antibody | Activated Caspase-3 (Apoptosis) | Highly specific marker for cells committed to apoptotic death. | Detects the proteolytically cleaved, active form of caspase-3, a key executioner caspase. |
| GFP-LC3 Plasmid | Autophagosomes (Autophagy) | Live-cell reporter; GFP-LC3 redistributes from diffuse to punctate structures upon autophagy induction. | Puncta represent autophagosomes; can be quantified by fluorescence microscopy or flow cytometry [69]. |
Integrated Experimental Workflow for Specific Distinction
A sequential, multi-parametric approach is recommended to conclusively identify the dominant cell death pathway.
Diagram: Decision Workflow for RCD Identification. A stepwise protocol integrating morphological, biochemical, and functional assays to distinguish between apoptosis, autophagy, and necroptosis with high specificity.
This integrated strategy, combining the gold standard of TEM morphology with specific biochemical assays and pharmacological inhibition, provides a robust framework for researchers to accurately delineate cell death mechanisms, thereby ensuring the specificity and validity of their conclusions.
The accurate identification of early apoptosis via transmission electron microscopy (TEM) is a cornerstone of cellular biology and drug development research. However, a one-size-fits-all protocol is a frequent pitfall that can compromise data integrity. The inherent biological diversity across cell types—from their structural components to their death pathway preferences—demands a tailored experimental approach. This guide provides a structured framework for researchers to optimize apoptosis induction and detection protocols for specific experimental models, ensuring high-fidelity morphological data. Recognizing that a protocol optimized for epithelial cells may fail for immune cells is the first step toward obtaining reliable, reproducible results in transmission electron microscopy identification of early apoptosis research [59] [17].
The foundation of any robust protocol is understanding the source of cell-type-specific variations. These include differences in the expression of key regulatory proteins (e.g., caspases, Bcl-2 family members), baseline metabolic activity, adhesion properties, and the inherent dominance of the intrinsic or extrinsic apoptotic pathway [14]. Furthermore, as highlighted by advanced detection systems like ADeS, the morphological hallmarks of apoptosis itself, such as nuclear condensation in epithelial cells versus membrane blebbing in leukocytes, are expressed differently across cell types, necessitating tailored recognition criteria [59]. This guide synthesizes current methodologies and quantitative data to empower scientists to make informed decisions for their specific experimental context.
Cell-Type-Specific Apoptotic Hallmarks and Detection Signatures
The morphological and biochemical events of apoptosis, while following a conserved sequence, manifest with significant variation across different cell types. Intravital microscopy and deep learning studies have quantitatively demonstrated that these differences are not merely anecdotal but are systematic and measurable [59]. For instance, the process of membrane blebbing and the formation of apoptotic bodies can differ in timing, scale, and specific structure.
A key discovery is the "FOotprint Of Death" (FOOD), a mechanism for generating large, substrate-bound extracellular vesicles during apoptosis. This phenomenon has been observed across a wide range of cell types, including human squamous epithelial cells (A431), primary human umbilical vein endothelial cells (HUVECs), mouse embryonic fibroblasts (MEFs), and human cervical adenocarcinoma (HeLa) cells [17]. The consistency of FOOD formation, occurring in ~80-99% of apoptotic cells across these types, suggests it is a fundamental process. However, its specific characteristics, such as the number of membranous branches and the area occupied, can vary, which may influence how cell death is identified and quantified in different models [17].
The following table summarizes key apoptotic features and their cell-type-specific expressions, which are critical for accurate TEM identification.
Table 1: Cell-Type-Specific Variations in Apoptotic Features Relevant for TEM Identification
| Cell Type | Nuclear Morphology | Cytoplasmic & Membrane Events | Key Biochemical Markers | TEM Identification Tips |
|---|---|---|---|---|
| Epithelial Cells (e.g., A431, HeLa) | Prominent chromatin condensation and nuclear fragmentation [59]. | Formation of FOOD, generating large substrate-bound vesicles [17]. | Caspase-3 activation, PARP cleavage [70]. | Focus on nuclear disintegration and vesicle formation at adhesion sites. |
| Leukocytes (e.g., Neutrophils) | Less pronounced nuclear condensation. | Dominant membrane blebbing and formation of apoptotic bodies in suspension [59]. | Phosphatidylserine (PS) externalization (Annexin V binding) [71]. | Look for membrane blebs and apoptotic bodies in the extracellular space. |
| Fibroblasts (e.g., MEFs) | Standard chromatin condensation. | Robust FOOD formation; may lack apoptopodia, reducing classical apoptotic body formation [17]. | Cytochrome c release, caspase-9 activation [14]. | Search for extensive membranous footprints on the substrate. |
| Neurons | Condensation in synaptic terminals and cell body. | Synaptic loss, degeneration of processes. | Active caspase-3, phosphorylated tau. | Examine synapses and axons for early signs of dismantling. |
Deep Learning Validation of Morphological Diversity
The development of the ADeS (Apoptosis Detection System) deep learning tool further validates the necessity of cell-type-optimized models. ADeS was specifically trained on two distinct datasets: one of epithelial cells and another of leukocytes, which involved different imaging modalities and staining techniques [59]. The model achieved over 98% classification accuracy by learning the unique morphological hallmarks that define apoptosis in each cell type. For the in vitro epithelial model, the defining features were nuclear shrinkage and chromatin condensation observed via nuclear markers. In contrast, for the in vivo leukocyte model, the key features were membrane blebbing and the formation of apoptotic bodies highlighted by cytoplasmic and membrane staining [59]. This underscores that effective detection, whether by AI or human analysis, depends on a foundational knowledge of cell-type-specific morphology.
Optimized Experimental Protocols for Apoptosis Induction and Analysis
Protocol for Specific Death Pathway Induction in Murine Tumors
For research requiring precise control over the cell death pathway, an inducible dimerizer system in mouse tumor models provides a method to trigger "pure" apoptosis or necroptosis. This is particularly valuable for studying the distinct morphological outcomes of each pathway in TEM.
Key Steps:
- Cell Line Generation: Lentivirally transduce your cell line of interest with constructs that allow for inducible activation of key apoptotic (e.g., caspases) or necroptotic (e.g., RIPK3/MLKL) executers [72] [73].
- Tumor Establishment: Implant the engineered cells into suitable murine models to establish tumors.
- Death Induction: Administer the specific dimerizer drug to trigger the designated death pathway (apoptosis or necroptosis) within the established tumor [72].
- Validation and Analysis: Use flow cytometry to optimize the death induction schedule and validate pathway specificity via Western blot for markers like cleaved caspase-3 (apoptosis) or phosphorylated MLKL (necroptosis) [72] [14]. Tissue samples can then be processed for TEM to compare the ultrastructural differences between the two death modes.
Automated, Label-Free Detection of Apoptosis in Co-culture Assays
In complex environments like tumor-immune cell co-cultures, label-free detection of apoptosis based on morphology offers a significant advantage by minimizing cellular perturbation.
Key Steps:
- Setup: Culture effector (e.g., T cells) and target (e.g., melanoma cells) in nanowell arrays. For identification, cells can be pre-labeled with fluorescent cytoplasmic dyes (e.g., PKH67 for T cells, PKH26 for tumor cells) [74].
- Imaging: Acquire time-lapse phase-contrast and fluorescent images, for example, every 5 minutes using a high-throughput microscope [74].
- Analysis with Deep Learning:
- Apoptotic Body Detection: Apply a trained ResNet50 convolutional neural network to identify nanowells that contain apoptotic bodies (ApoBDs) from the phase-contrast images. This method has been shown to achieve 92% accuracy [74].
- Onset Prediction: Use a temporal constraint (e.g., detection in three consecutive frames) to pinpoint the onset of apoptosis, reducing false positives.
- Correlation: This label-free method has been demonstrated to detect a significant number of apoptosis events (70%) that were not concurrently detected by Annexin-V staining, suggesting it may identify earlier events [74]. Correlate phase-contrast and fluorescent data for validation before moving to TEM sampling of specific time points.
Western Blot Analysis for Biochemical Confirmation
Western blotting remains a essential complementary technique to TEM for confirming the biochemical activation of apoptotic pathways.
Key Steps:
- Sample Preparation: Lyse cells or tissues and quantify protein concentration to ensure equal loading [70].
- Separation and Transfer: Separate proteins by SDS-PAGE and transfer to a membrane.
- Antibody Probing:
- Use antibodies against key apoptotic markers:
- Efficiency Tip: Use pre-mixed apoptosis antibody cocktails to detect multiple markers simultaneously, saving time and reagents while improving reproducibility [70].
- Quantification: Normalize band intensities to a housekeeping protein (e.g., β-actin, GAPDH). Use densitometry software (e.g., ImageJ) to calculate the ratio of cleaved to full-length protein, which indicates the level of apoptotic activation [70].
Visualizing Apoptotic Pathways and Workflows
Understanding the biochemical pathways and experimental workflows is crucial for protocol optimization. The following diagrams illustrate the core intrinsic apoptotic pathway and a generalized strategy for optimizing detection protocols.
The Intrinsic Apoptotic Pathway
The intrinsic (mitochondrial) pathway is a central mechanism of apoptosis initiated by internal cellular stress. The diagram below outlines the key molecular events.
Diagram Title: Intrinsic Apoptosis Pathway
Protocol Optimization Workflow
A systematic approach is required to tailor apoptosis detection for a new or unfamiliar cell type. The following workflow provides a logical sequence for this optimization.
Diagram Title: Protocol Optimization Workflow
The Scientist's Toolkit: Essential Reagents and Materials
Successful optimization relies on a core set of high-quality reagents and materials. The table below lists key solutions used in the protocols cited in this guide.
Table 2: Research Reagent Solutions for Apoptosis Detection
| Reagent/Material | Function | Example Application |
|---|---|---|
| Inducible Dimerizer System | Allows precise, specific induction of apoptosis or necroptosis in engineered cell lines [72] [73]. | Studying pure death pathways in murine tumor models. |
| BH3 Mimetics (e.g., ABT-737) | Small molecules that inhibit anti-apoptotic Bcl-2 proteins to induce intrinsic apoptosis [17]. | Specific induction of the mitochondrial apoptosis pathway. |
| Annexin V (FITC/647 Conjugates) | Binds to externalized phosphatidylserine (PS), a marker of early apoptosis. Used with viability dyes (PI) [74] [71]. | Flow cytometry or fluorescence microscopy to detect early apoptosis. |
| Apoptosis Antibody Cocktails | Pre-mixed antibodies for multiple markers (e.g., caspase-3, PARP, Bcl-2). Streamlines Western blot workflow [70]. | Efficient biochemical confirmation of apoptosis. |
| Caspase Substrate Peptides (e.g., DEVD) | Peptide sequences (DEVD for caspase-3) used in electrochemical biosensors or fluorescent assays [75]. | Sensitive detection of caspase enzyme activity. |
| JC-1 Dye | Lipophilic cationic dye used as a potentiometric probe for detecting loss of mitochondrial membrane potential (ΔΨm) [71]. | Flow cytometry or fluorescence microscopy to detect early apoptotic event. |
| Nanowell Arrays (e.g., PDMS) | Microwell chips for high-throughput, time-lapse imaging of single-cell and cell-cell interactions [74]. | Label-free apoptosis detection in co-cultures. |
Transmission Electron Microscopy (TEM) is an indispensable tool in cell biology, providing the nanometer-scale resolution necessary to visualize the subtle ultrastructural changes that characterize early apoptosis. Consistent and reproducible TEM analysis is therefore paramount for generating reliable data in foundational apoptosis research and drug development. This guide outlines a comprehensive quality control framework to achieve this goal, specifically within the context of identifying early apoptotic events.
Core Principles of TEM Operation for Apoptosis Research
Understanding and controlling key TEM parameters is the first step toward reproducible imaging. The configuration of the electron beam directly impacts image quality and the interpretation of cellular morphology.
Optimizing Key Imaging Parameters
The following parameters must be calibrated for each sample to balance resolution, contrast, and sample integrity.
| Parameter | Typical Setting for Biological Samples | Impact on Image Quality & Reproducibility | Rationale in Apoptosis Context |
|---|---|---|---|
| Acceleration Voltage [76] | 60-80 kV | Lower voltage increases contrast but reduces penetration; higher voltage reduces contrast but improves penetration. | Optimal contrast for visualizing early membrane blebbing and chromatin condensation without damaging cellular ultrastructure. |
| Beam Current [77] | Low to Moderate | High current improves signal-to-noise but can cause charging and sample damage; low current requires longer dwell times. | Preuces beam-induced damage to delicate apoptotic cells, preserving the integrity of early morphological signs like organelle swelling. |
| Pixel Dwell Time [77] | Optimized for SNR | Longer dwell time improves signal-to-noise ratio but increases total acquisition time and potential drift. | Enables clear visualization of low-contrast features, such as the disintegration of the nuclear envelope, with minimal noise. |
Automation in modern TEM systems, including automated alignment and calibration, further reduces operational variability and the risk of data distortion, ensuring that observations of apoptotic markers are consistent across sessions and operators [76].
A Standardized Workflow for Apoptosis-Focused TEM Analysis
A rigorous, step-by-step protocol is essential to minimize artifacts and ensure that observed structures are biologically relevant. The workflow below integrates quality control at every stage.
Detailed Experimental Protocols
Sample Preparation and Staining for Apoptotic Cells
- Cell Pellet Fixation: After inducing apoptosis, pellet cells and fix in a mixture of 2.5% glutaraldehyde and 2% paraformaldehyde in a 0.1 M cacodylate buffer (pH 7.2) for at least 1 hour at room temperature [78]. Post-fix with 1% osmium tetroxide for 1 hour to enhance membrane contrast [78].
- Dehydration and Embedding: Dehydrate the fixed pellet through a graded series of ethanol (e.g., 50%, 70%, 90%, 100%) and embed in a resin, such as Epon 812 [78].
- Ultramicrotomy and Staining: Use an ultramicrotome (e.g., Leica Ultracut UCT) to cut thin sections of 60-90 nm [78]. Collect sections on TEM grids and stain with 2% uranyl acetate followed by Reynolds lead citrate to provide contrast for chromatin and organelles [78].
Image Acquisition and Analysis
- Microscope Calibration: Follow the standardized workflow above. Use atomic-scale image calibration features to ensure accurate magnification [76].
- Blinded Analysis: To prevent bias, images should be coded and analyzed by researchers blinded to the experimental conditions. Multiple independent analysts should assess each image for key apoptotic markers to establish inter-observer reliability.
Correlative Analysis: Integrating TEM with Apoptosis Biomarkers
TEM provides ultrastructural "ground truth," but its power is magnified when correlated with biochemical assays. This multi-modal approach validates findings and links morphology to molecular pathways.
Key Apoptosis Markers for Correlative Assays
| Marker Category | Specific Marker | Function & Role in Apoptosis | Detectable Change |
|---|---|---|---|
| Nuclear Protein | PARP-1 (Poly (ADP-ribose) polymerase 1) [70] | DNA repair enzyme. Cleaved by executioner caspases. | Cleavage: Full-length (116 kDa) → Cleaved fragment (89 kDa). Presence confirms caspase activation. |
| Executioner Caspase | Caspase-3 [70] | Key protease that dismantles the cell. | Activation: Pro-caspase (inactive) → cleaved caspase-3 (active). Primary indicator of execution phase. |
| Regulatory Protein | Bcl-2 [70] | Anti-apoptotic protein. Maintains mitochondrial integrity. | Phosphorylation: Change in phosphorylation status can inactivate Bcl-2, promoting apoptosis. |
Western blot analysis for these markers provides a quantitative measure of apoptosis that complements qualitative TEM images. The signal intensity of cleaved proteins (e.g., cleaved caspase-3) should be normalized to the total protein levels and a housekeeping protein (e.g., β-actin) for accurate quantification [70].
The Apoptotic Signaling Pathway
The intrinsic apoptotic pathway, a common trigger, leads to the distinctive morphological changes visible via TEM. The following diagram illustrates this cascade.
The stages of nuclear condensation—Stage 1 (ring condensation), Stage 2 (necklace condensation), and Stage 3 (nuclear collapse/disassembly)—represent a defined biochemical program that can be precisely mapped using TEM [78]. It is important to note that Stage 1 ring condensation can occur independently of DNase activity, but subsequent stages require it, while ATP hydrolysis is specifically required for the final nuclear disassembly [78].
The Scientist's Toolkit: Essential Research Reagents and Materials
A selection of key reagents and materials critical for preparing and analyzing apoptotic samples via TEM is listed below.
| Item | Function in TEM Apoptosis Analysis |
|---|---|
| Glutaraldehyde/Paraformaldehyde | Primary fixative that cross-links proteins, stabilizing cellular morphology at the moment of fixation. |
| Osmium Tetroxide | Post-fixative that stabilizes lipids and provides electron density to membranes, crucial for visualizing membrane blebbing. |
| Uranyl Acetate & Lead Citrate | Heavy metal stains that bind to cellular components (e.g., DNA, membranes), enhancing contrast for clear imaging. |
| Epon 812 or Equivalent Resin | Embedding medium that infiltrates the tissue, allowing for the cutting of thin, stable sections for the electron beam. |
| Antibodies for Caspase-3 & Cleaved PARP | Used in western blotting to biochemically confirm the activation of apoptotic pathways correlated with TEM images [70]. |
| Apoptosis-Inducing Agents (e.g., Staurosporine) | Positive controls used to validate the entire TEM and biochemical workflow by reliably inducing apoptosis. |
Beyond Morphology: Validating TEM Findings with Complementary Biochemical and Functional Assays
The accurate identification of early apoptosis is crucial in diverse fields of biomedical research, including cancer biology, toxicology, and drug development. Individually, the gold-standard morphological assessment provided by transmission electron microscopy (TEM), the early-stage detection capability of Annexin V/Propidium Iodide (PI) staining, and the functional confirmation from caspase activation assays each provide distinct yet incomplete insights into the cell death process [79] [14] [80]. This technical guide details a robust integrated methodology that correlates these three powerful techniques, enabling researchers to achieve a comprehensive, multi-parameter validation of apoptotic events from initial triggering to final execution. This approach is particularly valuable for confirming the mechanism of action of novel therapeutic compounds, where distinguishing between apoptotic and non-apoptotic programmed cell death is functionally significant [81] [82].
Core Principles of Individual Techniques
Transmission Electron Microscopy (TEM) for Apoptosis
TEM provides the highest resolution for identifying the characteristic ultrastructural hallmarks of apoptosis, which remain the definitive standard for its diagnosis [29]. During apoptosis, cells undergo a series of distinctive morphological changes observable via TEM.
- Early Stage: The earliest signs include cell shrinkage, chromatin condensation (pyknosis), and compaction into dense masses that often aggregate at the nuclear periphery [1] [29].
- Mid Stage: The nucleus fragments (karyorrhexis), and the cell displays extensive plasma membrane blebbing while maintaining organelle integrity [1] [14].
- Late Stage: The cell disintegrates into multiple, discrete, membrane-bound apoptotic bodies, which contain tightly packed cytoplasm and organelles. These bodies are subsequently phagocytosed by neighboring cells without eliciting an inflammatory response [1] [29].
Critically, TEM can distinguish apoptosis from other death modalities like necrosis, which features cell swelling and membrane rupture, or autosis, characterized by pronounced ballooning of the perinuclear space [82].
Annexin V/Propidium Iodide (PI) Staining
This flow cytometry-based assay detects biochemical alterations in the plasma membrane, serving as a marker for the early and intermediate stages of apoptosis.
- Phosphatidylserine (PS) Externalization: In viable cells, PS is restricted to the inner leaflet of the plasma membrane. Early in apoptosis, PS is translocated to the outer leaflet, where it can be bound by fluorescently conjugated Annexin V (e.g., Annexin V-FITC) [80].
- Membrane Integrity Assessment: Propidium Iodide (PI) is a DNA-binding dye that is excluded from cells with an intact plasma membrane. Its incorporation indicates a loss of membrane integrity, a feature of late apoptosis and necrosis [80].
The resulting staining profile allows for the discrimination of cell populations:
- Annexin V–/PI–: Viable, healthy cells.
- Annexin V+/PI–: Early apoptotic cells.
- Annexin V+/PI+: Late apoptotic cells.
- Annexin V–/PI+: Necrotic cells or cellular debris.
Caspase Activation Assays
Caspases, a family of cysteine-aspartic proteases, are the central executioners of apoptosis. Their activation signifies an irreversible commitment to cell death [79] [14]. Detection methods include:
- Western Blotting: Identifies the cleavage of inert procaspases into their active subunits or the cleavage of canonical caspase substrates, such as Poly (ADP-ribose) Polymerase (PARP). Cleaved PARP is a widely used biochemical marker for apoptosis [80].
- Fluorescent Assay Kits: Utilize substrates that become fluorescent upon cleavage by active caspases (e.g., Caspase-3/7), allowing for quantification of activity in cell populations [83].
- Immunofluorescence: Can visualize the presence and localization of active caspases within fixed cells.
Caspase-3 and Caspase-7 are key "executioner" caspases, while Caspase-8 and Caspase-9 are initiators of the extrinsic and intrinsic pathways, respectively [79] [14].
Integrated Experimental Workflow
The following diagram illustrates the sequential and correlative workflow for the integrated detection of apoptosis.
Detailed Methodologies
Sample Preparation for Correlative Analysis
To ensure meaningful correlation, it is imperative to treat and harvest cell samples under identical conditions for all three assays. A time-course experiment following the application of an apoptotic stimulus (e.g., a chemotherapeutic agent) is highly recommended to capture the dynamic progression of cell death.
Cell Culture and Treatment:
- Culture cells (e.g., HCT116 colorectal cancer cells or other relevant models) according to standard protocols [81].
- Apply the apoptotic stimulus to treatment groups, while control groups receive vehicle only.
- Harvest cells at predetermined time points (e.g., 0, 6, 12, 24, 48 hours) post-treatment.
Annexin V/PI Staining Protocol
This protocol is adapted for flow cytometry analysis [80].
- Harvesting: Gently collect both adherent and floating cells to capture the entire population. Use trypsin without EDTA where possible, as EDTA can affect Annexin V binding.
- Washing: Wash cells twice with cold Phosphate-Buffered Saline (PBS).
- Binding: Resuspend the cell pellet (1-5 x 10⁵ cells) in 100 µL of 1X Binding Buffer.
- Staining: Add 5 µL of Annexin V-FITC and 5 µL of Propidium Iodide (PI) solution to the cell suspension.
- Incubation: Incubate for 15 minutes at room temperature (25°C) in the dark.
- Analysis: Within 1 hour, add 400 µL of 1X Binding Buffer to each tube and analyze by flow cytometry. Use FITC (FL1) and PI (FL2 or FL3) channels for detection.
Transmission Electron Microscopy (TEM) Sample Preparation
This protocol outlines the standard chemical fixation process for cultured cells [29].
- Primary Fixation: Pellet cells and fix in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for at least 1 hour at 4°C.
- Washing: Wash cells 3 times in 0.1 M cacodylate buffer, 10 minutes each.
- Post-Fixation: Post-fix in 1% osmium tetroxide in the same buffer for 1 hour at 4°C.
- Dehydration: Dehydrate through a graded ethanol series (50%, 70%, 90%, 100%) and finally with propylene oxide.
- Embedding: Infiltrate and embed cells in Epoxy resin (e.g., Epon or Araldite).
- Sectioning and Staining: Polymerize resin blocks, cut ultrathin sections (60-90 nm), and stain with uranyl acetate and lead citrate.
- Imaging: Examine sections using a TEM operating at 60-80 kV. Capture images of multiple random fields for quantitative assessment.
Caspase Activation Assay
A Western blot protocol for detecting caspase-3 and its substrate PARP is described below [80].
- Cell Lysis: Lyse harvested cells in RIPA buffer supplemented with protease and phosphatase inhibitors.
- Protein Quantification: Determine protein concentration using a BCA or Bradford assay.
- Gel Electrophoresis: Separate 20-30 µg of total protein per lane by SDS-PAGE.
- Transfer: Transfer proteins from the gel to a PVDF or nitrocellulose membrane.
- Blocking: Block the membrane with 5% non-fat milk in TBST for 1 hour.
- Antibody Incubation:
- Incubate with primary antibodies (e.g., anti-Caspase-3, anti-cleaved Caspase-3, anti-PARP, and a loading control like anti-GAPDH) overnight at 4°C.
- Wash the membrane and incubate with an appropriate HRP-conjugated secondary antibody for 1 hour.
- Detection: Develop the blot using a chemiluminescent substrate and image with a digital system.
Data Interpretation and Correlation
Successful integration relies on correlating data from all three techniques to build a coherent timeline of apoptotic events.
Expected Morphological and Biochemical Timeline
The following table summarizes the expected correlative results across the apoptotic timeline.
Table 1: Correlative Timeline of Apoptotic Markers
| Apoptotic Stage | TEM Morphology | Annexin V/PI Staining | Caspase Activation |
|---|---|---|---|
| Early | Chromatin condensation (pyknosis); cell shrinkage; intact organelles. | Annexin V+/PI– | Procaspase cleavage initiated; substrate (e.g., PARP) cleavage begins. |
| Mid | Nuclear fragmentation (karyorrhexis); pronounced membrane blebbing. | Annexin V+/PI– transitioning to Annexin V+/PI+ | High levels of active caspase (e.g., Caspase-3); significant PARP cleavage. |
| Late | Formation of apoptotic bodies; phagocytosis by adjacent cells. | Predominantly Annexin V+/PI+ | Caspase activity may decrease; cleaved substrates are evident. |
Quantitative Data Integration
To facilitate comparison, quantitative data from each assay should be compiled. The table below provides an example of expected outcomes from a treated cancer cell model, such as HCT116 cells.
Table 2: Exemplar Quantitative Data from an Apoptosis Induction Experiment
| Experimental Group | % Apoptotic Cells (TEM) | % Annexin V+ Cells (Flow Cytometry) | Caspase-3/7 Activity (Fold Change) | PARP Cleavage (Densitometry) |
|---|---|---|---|---|
| Control (Vehicle) | 2.5 ± 0.5% | 4.1 ± 0.8% | 1.0 ± 0.2 | Baseline |
| Treatment (6h) | 15.3 ± 2.1% | 18.5 ± 3.2% | 3.5 ± 0.6 | 2.8 ± 0.4 |
| Treatment (24h) | 55.7 ± 4.8% | 62.3 ± 5.1% | 8.2 ± 1.1 | 6.5 ± 0.9 |
Interpretation Guide:
- A strong correlation between the percentage of apoptotic cells identified by TEM and the percentage of Annexin V+ cells by flow cytometry validates the early apoptotic phenotype.
- A significant increase in caspase activity and PARP cleavage at the 6-hour time point confirms the activation of the executioner mechanism, preceding the widespread morphological changes seen at 24 hours.
- Discrepancies, such as high Annexin V staining without subsequent caspase activation or classical morphology, may indicate alternative cell death pathways or assay-specific artifacts [82].
The Scientist's Toolkit: Essential Research Reagents
The following table lists key reagents and their critical functions in performing this integrated apoptosis analysis.
Table 3: Essential Research Reagents for Integrated Apoptosis Detection
| Reagent / Kit | Function / Application | Key Characteristics |
|---|---|---|
| Annexin V-FITC/PI Apoptosis Detection Kit | Flow cytometry-based detection of PS externalization and membrane integrity. | Typically includes Annexin V-FITC, PI, and binding buffer. Allows for live cell staining and rapid quantification. |
| Caspase-3/7 Activity Assay Kit | Luminescent or fluorescent measurement of executioner caspase activity. | Provides a cleavable substrate; results in a signal proportional to caspase activity in the sample. |
| Anti-Cleaved Caspase-3 Antibody | Immunodetection of activated caspase-3 via Western blot or immunofluorescence. | Highly specific for the large fragment of caspase-3 resulting from cleavage; confirms activation. |
| Anti-PARP Antibody | Immunodetection of full-length and cleaved PARP via Western blot. | Cleavage of PARP (from ~116 kDa to ~89 kDa) is a hallmark biochemical event in apoptosis. |
| Glutaraldehyde & Osmium Tetroxide | Primary and post-fixatives for TEM sample preparation. | Glutaraldehyde cross-links proteins; Osmium tetroxide fixes lipids and provides electron density. |
| Epoxy Resin (e.g., Epon 812) | Embedding medium for TEM samples. | Provides hard, stable blocks suitable for cutting ultrathin sections. |
Troubleshooting and Integration Guidelines
- Annexin V False Positives: Certain cell types or treatment conditions (e.g., mechanical stress) can cause nonspecific Annexin V binding. Always include untreated controls and correlate with other markers [80].
- TEM Artifacts: Improper fixation or dehydration can introduce artifacts like cytoplasmic vacuolization or shrinkage that may be mistaken for pathology. Adherence to protocol is critical [29].
- Caspase-Independent Apoptosis: In some instances, cell death may exhibit classical apoptotic morphology (as seen by TEM) without significant caspase activation. This highlights the necessity of morphological confirmation and may point to alternative death mechanisms [82].
- Definitive Identification: The presence of chromatin condensation and apoptotic bodies via TEM remains the definitive standard for confirming apoptosis. Positive Annexin V/PI staining and caspase activation should be interpreted as supportive evidence that, when correlated with morphology, provides a powerful and holistic confirmation of apoptotic cell death.
Within the context of a broader thesis on transmission electron microscopy (TEM) identification of early apoptosis, this technical guide provides a comparative analysis of TEM and light microscopy. The strategic manipulation of regulated cell death (RCD) pathways, including apoptosis, has emerged as a crucial component in effective anti-tumor immunity and drug development [84]. Accurate detection of apoptosis is therefore fundamental for researchers and scientists investigating cancer therapeutics, cardiovascular diseases, and neurodegenerative disorders. This whitepaper delineates the technical capabilities, methodological approaches, and practical applications of both TEM and light microscopy in identifying hallmark features of apoptotic cell death. We provide a detailed examination of their respective resolutions, diagnostic strengths, and limitations, supported by structured quantitative data, experimental protocols, and pathway visualizations to serve the needs of research professionals engaged in cell death studies.
Technical Comparison of Resolution and Diagnostic Capabilities
The fundamental difference between these microscopy techniques lies in their resolution limits and the type of information they yield. Light microscopy, including advanced forms like full-field optical coherence tomography (FF-OCT), offers live-cell imaging but is limited by the diffraction of light, typically achieving resolutions around 200 nm laterally [15]. In contrast, TEM utilizes a beam of electrons, providing sub-nanometer resolution and enabling the visualization of ultrastructural details within cells [85].
The following table summarizes the core technical parameters of each method for apoptosis detection:
Table 1: Technical Comparison of TEM and Light Microscopy for Apoptosis Detection
| Parameter | Transmission Electron Microscopy (TEM) | Light Microscopy |
|---|---|---|
| Resolution | Sub-nanometer level (e.g., ~0.2 nm) [85] | Diffraction-limited (~200 nm lateral) [15] |
| Key Apoptotic Features Visualized | Chromatin condensation, pyknotic nuclei, cytoplasmic vacuolization, degenerated organelles, mitochondrial swelling [85] | Cell shrinkage, membrane blebbing, nuclear fragmentation (with stains), caspase activation (with fluorescent reporters) [16] |
| Imaging Context | Static, high-detail ultrastructure | Dynamic, real-time processes in live cells |
| Sample Preparation | Complex (fixation, sectioning, staining) [85] | Simpler (can be label-free or with stains) [16] |
| Viability | Requires cell fixation (non-viable) | Suitable for live-cell imaging [16] |
| Cost & Complexity | High [16] | Moderate to high, depending on modality [16] |
| Real-time Monitoring | No | Yes [16] |
Light microscopy excels in real-time detection of apoptosis. Using transmitted light modalities like Differential Interference Contrast (DIC) or Phase Contrast (PC), researchers can quickly identify morphological changes such as cytoplasmic blebbing and cell shrinkage without staining [16]. Fluorescence light microscopy further enables visualization of specific apoptotic events using probes for DNA fragmentation (e.g., Hoechst), caspase activation (e.g., NucView 488), and membrane integrity (e.g., Annexin V) [16]. A novel fluorescent reporter technology that loses fluorescence upon caspase-3 cleavage has been developed for more sensitive and precise real-time monitoring of apoptosis [86].
TEM provides the definitive standard for confirming apoptosis through its ability to reveal ultrastructural pathology. Studies on oral squamous cell carcinoma (OSCC) cells treated with salivary exosomes used TEM to identify characteristic apoptotic features, including pyknotic nuclei (condensed, darkly staining nuclei) and cytoplasmic vacuolization, which were absent in untreated cells [85]. This level of detail is unobtainable with conventional light microscopy.
Advanced light microscopy techniques are bridging the resolution gap. Full-field optical coherence tomography (FF-OCT), a label-free, high-resolution interferometric technique, can visualize apoptotic features like echinoid spine formation, membrane blebbing, and filopodia reorganization at the single-cell level in real-time [15].
Table 2: Diagnostic Capabilities for Key Apoptotic Events
| Apoptotic Event | TEM Diagnostic Capability | Light Microscopy Diagnostic Capability |
|---|---|---|
| Chromatin Condensation | Direct visualization of highly condensed, marginalized chromatin [85] | Indirect via intense, punctate nuclear staining (e.g., with DAPI) [16] |
| Nuclear Fragmentation | Visualization of nuclear envelope breakdown and fragmented nuclei | Detection of multiple, discrete DNA-containing bodies via fluorescent stains [16] |
| Membrane Blebbing | Detailed view of bleb ultrastructure and plasma membrane integrity | Real-time observation of dynamic bleb formation and retraction [16] [15] |
| Mitochondrial Changes | Swelling, cristae breakdown, and release of contents [85] | Loss of mitochondrial membrane potential using fluorescent dyes (e.g., JC-1) [87] |
| Caspase Activation | Not directly visualized | Directly visualized with fluorescent activity reporters (e.g., for caspase-3/7) [16] [86] |
| Formation of Apoptotic Bodies | High-resolution view of content and membrane structure | Observation of released vesicles as phase-bright or fluorescent objects [16] |
Experimental Protocols for Apoptosis Detection
TEM Protocol for Apoptosis Identification
This protocol is adapted from studies on OSCC cells, detailing the steps for sample preparation, processing, and imaging to identify ultrastructural markers of apoptosis [85].
- Cell Culture and Fixation: Grow cells on appropriate substrates. Induce apoptosis as required. Perform primary fixation with 3% glutaraldehyde in a 0.1 M sodium cacodylate buffer (pH 7.0) for 2 hours at room temperature.
- Washing: Rinse the cells several times with the same 0.1 M sodium cacodylate buffer to remove excess glutaraldehyde.
- Secondary Fixation: Post-fix with 1% osmium tetroxide for 2 hours. This step stabilizes lipids and enhances membrane contrast.
- Dehydration: Gradually dehydrate the samples using an ascending ethanol series (e.g., from 10% to 100% ethanol), followed by a transition to absolute ethanol.
- Infiltration and Embedding: Infiltrate the cells with a resin, such as epoxy resin, using a resin/acetone mixture. Subsequently, embed the samples in pure resin and polymerize in an oven.
- Sectioning: Use an ultramicrotome to cut ultrathin sections (typically 60-90 nm thick) and mount them on copper grids.
- Staining: Stain the sections with heavy metal stains, such as uranyl acetate and lead citrate, to increase electron scattering and contrast.
- TEM Imaging: Examine the stained sections under the TEM operating at 80-100 kV. Identify and document key apoptotic features, including chromatin margination and condensation, pyknotic nuclei, cytoplasmic vacuolization, and degenerated organelles [85].
Light Microscopy Protocol for Real-Time Apoptosis Monitoring
This protocol leverages both label-free and fluorescence methods for detecting apoptosis in live cells, using reagents like staurosporine and NucView 488 [16].
- Cell Preparation and Plating: Culture cells (e.g., PtK or HeLa lines) in appropriate media. Plate cells onto glass-bottom Petri dishes suitable for high-resolution microscopy.
- Induction of Apoptosis: To induce intrinsic apoptosis, treat cells with 10 μM Staurosporine (in 1% DMSO) approximately 30 minutes prior to imaging [16].
- Staining for Caspase Activity (Optional): For fluorescent detection of executioner caspase activation, incubate cells with a reagent like the NucView 488 caspase-3/7 substrate according to the manufacturer's instructions. This non-fluorescent substrate penetrates the plasma membrane and is cleaved by active caspase-3/7, releasing a DNA-binding dye that stains the nucleus [16].
- Time-Lapse Imaging:
- Label-free Imaging: Use transmitted light modalities like DIC or Phase Contrast on an inverted microscope to capture morphological changes (cell shrinkage, blebbing) over time. Acquire images at a rate of 2-4 frames per minute.
- Fluorescence Imaging: Using the same microscope with fluorescent optics, capture images of the NucView 488 signal (excitation/emission ~488/520 nm) to monitor caspase activation.
- Environmental Control: Maintain cells at 37°C with 5% CO₂ throughout the imaging session to ensure viability.
- Data Analysis: Correlate the onset of morphological changes (from DIC) with the activation of caspases (from fluorescence) to confirm apoptosis and determine the sequence of events.
Visualizing the Apoptotic Pathway and Experimental Workflow
The core executioner of apoptosis is caspase-3. The following diagram illustrates the simplified intrinsic pathway and the principle behind a novel fluorescent reporter for its detection.
Caspase-3 Activation & Reporter Detection
The decision between using TEM or light microscopy depends on the research question. The following workflow outlines a logical approach for selecting and applying these techniques in an apoptosis detection experiment.
Apoptosis Detection Experimental Workflow
Essential Research Reagent Solutions
The following table compiles key reagents and materials used in the featured experiments for detecting apoptosis, along with their specific functions.
Table 3: Key Research Reagent Solutions for Apoptosis Detection
| Reagent / Material | Function / Application | Experimental Context |
|---|---|---|
| Staurosporine | Protein kinase inhibitor; induces intrinsic apoptosis through caspase-dependent and independent pathways [16]. | Used to experimentally induce apoptosis in mammalian cell lines (e.g., PtK, HeLa) for microscopy [16]. |
| NucView 488 | Fluorescent caspase-3/7 substrate. Becomes fluorescent upon cleavage, staining the nucleus [16]. | For real-time visualization of caspase activation in live cells using fluorescence microscopy [16]. |
| Annexin V | Binds to phosphatidylserine (PS), which is externalized to the outer leaflet of the plasma membrane during early apoptosis [16]. | Detection of mid-stage apoptosis via fluorescence microscopy or flow cytometry [16]. |
| Glutaraldehyde | Cross-linking fixative that stabilizes protein structure. | Primary fixative for TEM sample preparation to preserve ultrastructure [85]. |
| Osmium Tetroxide | Fixative and stain that stabilizes lipids and adds contrast to membranes. | Secondary fixative in TEM protocol [85]. |
| Uranyl Acetate & Lead Citrate | Heavy metal stains that scatter electrons. | Post-embedding staining of ultrathin sections to enhance contrast for TEM imaging [85]. |
| Novel GFP-based Reporter | Engineered fluorescent reporter that loses fluorescence upon caspase-3 cleavage at the DEVDG sequence [86]. | Highly sensitive and specific real-time apoptosis monitoring in human and animal cells [86]. |
| Doxorubicin | Chemotherapeutic agent that intercalates into DNA, causing double-strand breaks and inducing apoptosis [15]. | Used to induce apoptosis in cancer cell lines (e.g., HeLa) for imaging studies [15]. |
Both transmission electron microscopy and light microscopy provide indispensable, yet complementary, capabilities for apoptosis detection in biomedical research. TEM remains the unequivocal gold standard for confirming apoptosis through high-resolution visualization of definitive ultrastructural pathology, making it ideal for endpoint validation in thesis research. Light microscopy, particularly with advancements in real-time fluorescent reporters and label-free techniques like FF-OCT, offers unparalleled insight into the dynamics and kinetics of cell death in living systems. The choice between these techniques is not one of superiority but of strategic application. For a comprehensive understanding, from initial dynamic events to final ultrastructural confirmation, an integrated approach that leverages the strengths of both technologies is most powerful. This synergistic methodology provides researchers and drug development professionals with a complete toolkit for elucidating the mechanisms of cell death and evaluating novel therapeutic agents.
Transmission Electron Microscopy (TEM) stands as a powerful tool in cell biology, particularly for the precise identification of early apoptosis in research settings. Its unparalleled resolution allows scientists to visualize the initial ultrastructural changes that hallmark this form of programmed cell death. However, effectively leveraging TEM and choosing when to employ alternative methodologies requires a deep understanding of its specific capabilities and constraints. This guide provides researchers, scientists, and drug development professionals with a technical framework for integrating TEM into apoptosis studies, detailing optimal use cases, standardized protocols, and strategies for correlating TEM data with other functional assays to obtain a comprehensive biological picture.
The Role of TEM in Identifying Early Apoptosis
Key Advantages of TEM
TEM offers several distinct advantages for detecting early apoptotic events, primarily rooted in its high-resolution capabilities.
- Unmatched Resolution: TEM provides high spatial resolution down to 0.1 nm, enabling the visualization of single nanoparticles and their precise localization within cellular structures [27]. This level of detail is crucial for observing the subtle, early morphological changes in apoptosis.
- Definitive Morphological Identification: TEM is considered the most classic and reliable method for distinguishing between apoptotic and necrotic cell death based on distinct ultrastructural features [88]. It allows researchers to directly observe key hallmarks without inference.
Table 1: Key Ultrastructural Markers of Early Apoptosis Detectable by TEM
| Cellular Feature | Early Apoptotic Morphology | Significance |
|---|---|---|
| Chromatin | Condensation and margination along the nuclear envelope [88] | One of the earliest nuclear events |
| Cytoplasm | Shrinkage and compaction [88] | Indicates loss of water and ions |
| Plasma Membrane | Blebbing and preservation of integrity [88] [89] | Distinguishes apoptosis from necrosis; leads to apoptotic body formation |
| Organelles | Generally intact (e.g., mitochondria, ER) [90] | Contrasts with organellar swelling in necrosis |
Inherent Limitations and Challenges
Despite its strengths, TEM comes with inherent limitations that researchers must account for in their experimental design.
- Static and Two-Dimensional Imaging: Conventional TEM provides static, two-dimensional snapshots of a dynamic process. This can lead to an incomplete understanding of the temporal sequence of apoptotic events [27].
- Complex and Artifact-Prone Sample Preparation: The multi-step preparation process involving chemical fixation, resin embedding, and heavy metal staining is tedious and can introduce artifacts, such as the denaturation of delicate cellular structures [27].
- Limited Field of View and Sample Throughput: The high magnification of TEM restricts the observable area at any one time, making it difficult to survey large cell populations. It is not a high-throughput technique and is poorly suited for statistical analysis or quantification without extensive sampling [27] [90].
- Inability to Directly Assess Biochemical Function: TEM reveals morphology but cannot directly confirm the biochemical activity central to apoptosis, such as caspase activation or phosphatidylserine externalization [88] [89].
When to Rely on TEM: Optimal Use Cases and Protocols
TEM is the method of choice in several specific scenarios within apoptosis research.
- Definitive Morphological Confirmation: When you need to unambiguously confirm that cell death is occurring via apoptosis and not another pathway like necrosis or pyroptosis, which have distinctly different ultrastructural presentations [89].
- Analysis of Subcellular Localization: When the research question involves understanding the interaction of a drug, nanoparticle, or protein with specific organelles during apoptosis, such as mitochondrial permeabilization or ER stress [27] [91].
- Studying Novel or Complex Cell Death Phenotypes: When observing a cell death process with unknown characteristics, TEM can provide the foundational morphological data to classify it or distinguish it from known pathways.
Detailed Experimental Protocol for TEM in Apoptosis
The following workflow is critical for generating reliable TEM data.
Workflow Description:
- Induce Apoptosis: Treat cells with the apoptotic stimulus (e.g., chemotherapeutic agent, UV irradiation, ligand).
- Primary Fixation: Rapidly fix cells with 2.5% glutaraldehyde in a 0.1 M sodium cacodylate buffer (pH 7.4) to preserve ultrastructure. This cross-links proteins and stabilizes the cell in its current state [88].
- Washing: Rinse cells several times in buffer to remove excess fixative.
- Post-Fixation: Treat cells with 1% osmium tetroxide for 1-2 hours. Osmium tetroxide acts as a secondary fixative and strongly binds to lipids, stabilizing membranes and providing electron density [88].
- Dehydration: Gradually dehydrate the sample using a graded series of ethanol (e.g., 50%, 70%, 90%, 100%) to remove all water.
- Resin Infiltration and Embedding: Infiltrate the sample with a liquid epoxy resin (e.g., Epon or Spurr's) which is then polymerized in an oven at ~60°C to form a hard block.
- Ultra-thin Sectioning: Use an ultramicrotome with a diamond or glass knife to cut sections 60-90 nm thick. These thin sections are mounted on TEM copper grids.
- Contrast Staining: Stain grids with uranyl acetate and Reynold's lead citrate to enhance the scattering of electrons and improve contrast of cellular structures [88].
Advanced TEM Techniques
- Cryo-Electron Microscopy (Cryo-EM): Techniques like plunge-freezing or high-pressure freezing immobilize cells in a near-native, vitreous state, avoiding chemical fixation artifacts. Cryo-electron tomography (cryo-ET) can then be used to determine 3D structures at nanometer resolution, providing detailed insight into the spatial organization of apoptotic machinery [27].
- Electron Tomography: This technique involves collecting a series of 2D TEM images at different tilt angles, which are computationally reconstructed into a 3D model. It is powerful for visualizing the 3D architecture of organelles like mitochondria and the ER during apoptosis [27] [90].
- Correlative Light and Electron Microscopy (CLEM): CLEM combines the dynamic, functional data from light microscopy (e.g., live-cell imaging of apoptosis) with the high-resolution structural context of TEM. Researchers can first locate and analyze a specific event like mitochondrial membrane potential loss using fluorescence, then examine the exact same cell at ultrastructural resolution with EM [27].
When to Use Alternative Methods
While TEM is powerful, many research questions require alternative or complementary techniques.
For Quantitative and High-Throughput Analysis
- Flow Cytometry: This is the preferred method for quantifying the percentage of apoptotic cells in a large population.
- Annexin V/PI Staining: Detects the externalization of phosphatidylserine (PS) on the outer leaflet of the plasma membrane, an early event in apoptosis. Annexin V-FITC binds to exposed PS, while propidium iodide (PI) indicates late apoptosis/necrosis by staining DNA in cells with compromised membranes [88]. This allows for the discrimination of live (Annexin V-/PI-), early apoptotic (Annexin V+/PI-), and late apoptotic/necrotic (Annexin V+/PI+) cells.
- Caspase Activity Assays: Uses fluorescently-labeled inhibitors of caspases (FLICA) or antibodies against active caspases to detect the activation of these key executioner enzymes.
For Assessing Biochemical Activity and Molecular Markers
- Western Blotting: Essential for confirming the biochemical hallmarks of apoptosis.
- Immunofluorescence (IF): Allows for the subcellular localization of apoptotic markers (e.g., cytochrome c release from mitochondria) within the context of a fluorescently labeled cell, though at a lower resolution than TEM [92] [91].
For Functional and Live-Cell Imaging
- Live-Cell Imaging: Using fluorescent dyes and time-lapse microscopy, researchers can track the dynamics of apoptosis in real time.
- MMP Assays: Lipophilic cationic dyes like JC-1 are used to detect the loss of mitochondrial membrane potential (ΔΨm), an early and irreversible step in intrinsic apoptosis. In healthy cells, JC-1 forms aggregates that emit red fluorescence; in apoptotic cells, it remains in a monomeric state emitting green fluorescence [88].
- Nuclear Staining: Fluorescent DNA-binding dyes like Hoechst 33342 or DAPI can be used to observe nuclear fragmentation and chromatin condensation in live or fixed cells, though with less clarity than TEM [88].
Table 2: Comparison of Apoptosis Detection Methods
| Method | Key Readout | Throughput | Key Advantage | Key Disadvantage |
|---|---|---|---|---|
| TEM | Ultrastructural morphology | Low | Definitive morphological identification; Highest resolution | Static; No quantification; Artifact potential |
| Flow Cytometry (Annexin V) | PS externalization | High | Quantification of populations; Distinguishes early/late stages | No morphological context |
| Western Blot | Protein cleavage/expression | Medium | Molecular-level confirmation; Standard technique | No single-cell data; Requires large cell numbers |
| Live-Cell Imaging | Real-time dynamics | Medium | Kinetic data; Functional assessment | Lower resolution; Photo-toxicity |
Integrated Experimental Strategy: A Practical Workflow
A robust apoptosis study often integrates multiple techniques. The diagram below illustrates a logical workflow for a comprehensive investigation.
Workflow Description:
- Initial Screening: Use high-throughput methods like flow cytometry (Annexin V) or cell viability assays (CCK-8) to quickly identify conditions that induce apoptosis.
- Quantification and Kinetics: Employ flow cytometry to quantify the percentage of cells in different stages of apoptosis and establish a time-course for the event.
- Molecular Confirmation: Use Western blotting to detect key apoptotic proteins like cleaved caspase-3 and cleaved PARP, confirming the biochemical pathway involved.
- Functional Assessment: Utilize live-cell imaging and functional probes (e.g., JC-1) to observe the dynamics of apoptosis, such as the precise timing of mitochondrial membrane potential collapse.
- Ultimate Morphological Validation: Finally, use TEM to provide the definitive, high-resolution visual evidence of apoptosis, confirming that the observed biochemical and functional data correlate with the expected ultrastructural pathology.
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Reagents for Apoptosis Detection
| Reagent / Kit | Primary Function | Key Application |
|---|---|---|
| Glutaraldehyde & Osmium Tetroxide | Chemical fixation and membrane stabilization | TEM sample preparation [88] |
| Annexin V-FITC / PI Apoptosis Kit | Detection of PS externalization and membrane integrity | Flow cytometry-based quantification of early/late apoptosis [88] |
| JC-1 Dye | Detection of mitochondrial membrane potential (ΔΨm) loss | Fluorometric and flow cytometric analysis of early intrinsic apoptosis [88] |
| Antibodies: Cleaved Caspase-3, Cleaved PARP | Detection of key apoptotic protein cleavages | Western blotting and immunofluorescence for molecular confirmation [88] |
| Hoechst 33342 / DAPI | DNA staining for nuclear morphology | Fluorescence microscopy to observe chromatin condensation [88] |
| Caspase Activity Assay Kits (e.g., FLICA) | Detection of active caspase enzymes | Spectrophotometric or fluorometric functional assays [89] |
Validation of cellular events, particularly the detection of programmed cell death, is a cornerstone of biomedical research. This guide provides an in-depth technical framework for validating early apoptosis, with a specific focus on applications in cancer research and neurodegenerative diseases. The content is structured to serve the needs of researchers and drug development professionals, offering detailed methodologies, analytical frameworks, and practical tools for accurate apoptosis identification and interpretation across disease contexts. The emphasis is on leveraging transmission electron microscopy (TEM) as a definitive validation tool while integrating it with complementary biochemical and spectroscopic techniques to provide a multi-parameter assessment of cell death.
Apoptosis Signaling Pathways: Core Mechanisms and Detection Landmarks
Apoptosis progresses through two primary signaling pathways that converge on a common execution phase. Understanding these pathways provides the molecular basis for developing detection strategies.
Extrinsic (Death Receptor) Pathway
The extrinsic pathway initiates when extracellular ligands bind to death receptors on the cell surface [66] [79]. This includes:
- FasL/FasR and TNF-α/TNFR1 interactions: Ligand binding induces receptor trimerization and formation of the Death-Inducing Signaling Complex (DISC).
- DISC composition: FAS-associated death domain (FADD) adaptor proteins recruit and activate procaspase-8 [79].
- Initiation cascade: Active caspase-8 directly cleaves and activates executioner caspases (caspase-3, -7), initiating the apoptotic cascade [66].
Intrinsic (Mitochondrial) Pathway
The intrinsic pathway triggers through intracellular stress signals, including DNA damage, oxidative stress, and endoplasmic reticulum stress [66] [79]. Key events include:
- BCL-2 family regulation: Pro-apoptotic proteins (Bax, Bak) undergo conformational changes, permeabilizing the mitochondrial outer membrane, while anti-apoptotic members (Bcl-2, Bcl-xL) inhibit this process [93] [66].
- Cytochrome c release: Mitochondrial membrane permeability enables cytochrome c release into the cytosol, where it binds Apoptotic Protease-Activating Factor-1 (APAF-1) [79].
- Apoptosome formation: The cytochrome c/APAF-1 complex forms a heptameric apoptosome, recruiting and activating procaspase-9, which then activates executioner caspases [66].
Both pathways converge to activate executioner caspases (primarily caspase-3), leading to systematic cleavage of cellular substrates and characteristic morphological changes [93] [66].
Pathway Interconnectivity and Alternative Cell Death Modalities
Cross-talk occurs between pathways; caspase-8 cleaves Bid to tBid, which amplifies apoptosis through the mitochondrial pathway [79]. Additionally, other programmed cell death forms like necroptosis (RIPK1/RIPK3/MLKL-mediated) and pyroptosis (caspase-1/inflammasome-mediated) represent distinct but potentially overlapping pathways that may be activated in specific disease contexts or in response to therapeutic agents [66].
Figure 1: Apoptosis Signaling Pathways. This diagram illustrates the major extrinsic (red) and intrinsic (blue) apoptotic pathways, their convergence on executioner caspases (yellow), and the resulting morphological changes (green).
Technical Guide to Early Apoptosis Detection
Transmission Electron Microscopy for Early Apoptosis Identification
TEM provides the highest resolution morphological evidence for early apoptosis, serving as a gold standard for validating other detection methods [79]. The procedure requires careful sample preparation and interpretation.
Sample Preparation Protocol for TEM Apoptosis Analysis
- Primary Fixation: Fix cell pellets or tissue sections (1mm³) in 2.5% glutaraldehyde in 0.1M sodium cacodylate buffer (pH 7.4) for 2-4 hours at 4°C.
- Washing: Rinse 3 times in 0.1M sodium cacodylate buffer (15 minutes each).
- Post-fixation: Treat with 1% osmium tetroxide in 0.1M sodium cacodylate buffer for 1-2 hours at 4°C.
- Dehydration: Sequential ethanol series (30%, 50%, 70%, 90%, 100%) for 15 minutes each, followed by 100% acetone for 20 minutes.
- Infiltration and Embedding: Gradually infiltrate with epoxy resin (25%, 50%, 75%, 100%) over 8-12 hours, then embed in fresh resin and polymerize at 60°C for 48 hours.
- Sectioning: Cut ultrathin sections (60-90nm) using an ultramicrotome and collect on copper grids.
- Staining: Stain with uranyl acetate (10-15 minutes) and lead citrate (5-10 minutes) to enhance contrast.
- Imaging: Examine grids using TEM at 80kV, capturing images at various magnifications (2,000X to 30,000X).
Key Morphological Indicators of Early Apoptosis Accessible via TEM
During early apoptosis, TEM reveals:
- Chromatin condensation: Marginalization of chromatin against the nuclear envelope in dense, compact masses [66] [79].
- Nuclear membrane blebbing: Protrusions of the nuclear envelope containing condensed chromatin.
- Cytoplasmic condensation: Increased electron density of the cytosol with organelle compaction.
- Preserved organelle integrity: Mitochondria, Golgi apparatus, and endoplasmic reticulum remain largely intact initially.
- Plasma membrane blebbing: Formation of surface protrusions while membrane integrity remains intact.
Late apoptosis features include nuclear fragmentation (karyorrhexis), formation of membrane-bound apoptotic bodies containing organelles and nuclear fragments, and eventual secondary necrosis with loss of membrane integrity [93] [66].
TEM Image Processing and Analysis
- Contrast Enhancement: Use ImageJ/FIJI software (Process > Filters > Mean) with radius 0.5-3.0 pixels to reduce noise while preserving structural details [94].
- Brightness/Contrast Adjustment: Navigate to Image > Adjust > Brightness/Contrast to optimize visualization of ultrastructural features without altering underlying data [94].
- Scale Bar Addition: Apply Analyze > Set Scale using the specific pixel size for the image magnification, then add scale bars via Analyze > Tools > Scale Bar [94].
- Batch Processing: Implement macro scripts in ImageJ to ensure consistent application of filtering and contrast adjustments across all images from an experiment [94].
Complementary Biochemical and Cytometric Assays
Integrating TEM with biochemical methods provides comprehensive apoptosis validation through multiple detection parameters.
Phosphatidylserine Externalization Detection
The annexin V assay detects phosphatidylserine (PS) translocation from the inner to outer leaflet of the plasma membrane, an early apoptotic event [93].
Detailed Protocol:
- Harvest cells (5×10⁵) by gentle centrifugation (300 × g for 5 minutes).
- Wash twice in cold PBS and resuspend in 1X binding buffer.
- Add fluorochrome-conjugated annexin V (e.g., FITC, PE, BV421) and incubate for 15 minutes in the dark at room temperature.
- Add viability dye (e.g., 7-AAD, propidium iodide) 5 minutes before analysis to exclude necrotic cells.
- Analyze by flow cytometry within 1 hour using appropriate laser lines and detection filters.
- Interpretation: Annexin V+/viability dye- indicates early apoptosis; Annexin V+/viability dye+ indicates late apoptosis/secondary necrosis [93].
Caspase Activation Assessment
Caspase activity serves as a central biochemical indicator of apoptosis commitment.
Active Caspase Immunodetection:
- Fix and permeabilize cells using commercial fixation/permeabilization buffers.
- Incubate with antibodies specific for active caspase-3 (cleaved form) for 30-60 minutes at room temperature.
- For flow cytometry, use fluorochrome-conjugated secondary antibodies or direct conjugates.
- Analyze by flow cytometry, imaging, or western blot [93].
Fluorogenic Substrate Assay:
- Prepare cell lysates or incubate with cell-permeable substrates (e.g., PhiPhiLux, NucView 488).
- For live-cell analysis, incubate intact, unfixed cells with caspase substrates for 30-60 minutes.
- Measure fluorescence emission (caspase-3/7: Ex/Em ~488/530nm; caspase-8: Ex/Em ~488/530nm; caspase-9: Ex/Em ~488/530nm) [93].
Mitochondrial Membrane Potential (ΔΨm) Assessment
JC-1 Staining Protocol:
- Incubate cells (5×10⁵/mL) with 2μM JC-1 for 15-30 minutes at 37°C.
- Wash twice with PBS and analyze by flow cytometry or fluorescence microscopy.
- Interpretation: Healthy mitochondria with high ΔΨm form J-aggregates (red fluorescence, ~590nm); apoptotic cells with diminished ΔΨm maintain JC-1 as monomers (green fluorescence, ~529nm) [93].
DNA Fragmentation Analysis
TUNEL (TdT dUTP Nick-End Labeling) Assay:
- Fix cells in 1% paraformaldehyde for 15 minutes, then in 70% ethanol for at least 2 hours.
- Permeabilize with 0.1% Triton X-100 for 5 minutes.
- Incubate with TdT enzyme and fluorochrome-labeled dUTP (e.g., Br-dUTP) for 60 minutes at 37°C.
- Counterstain with DNA dye (e.g., 7-AAD, PI) for cell cycle analysis if desired.
- Analyze by flow cytometry or fluorescence microscopy [93].
Table 1: Apoptosis Detection Methods Comparison
| Detection Method | Target/Principle | Stage Detected | Key Advantages | Limitations |
|---|---|---|---|---|
| TEM Morphology | Ultrastructural changes | Early to late | Gold standard, high resolution | End-point, technical expertise required |
| Annexin V Binding | PS externalization | Early | Live cell application, quantitative | Cannot use with serum-containing media |
| Caspase Activation | Caspase cleavage/activity | Early to mid | High specificity, multiple formats | Transient signal, pathway-specific |
| ΔΨm Loss (JC-1) | Mitochondrial membrane potential | Early-mid | Functional assessment, live cells | Affected by metabolic inhibitors |
| TUNEL Assay | DNA fragmentation | Late | Specific for late apoptosis, histology compatible | Cannot detect early apoptosis |
| Cellular Viability | Membrane integrity | Late apoptosis/necrosis | Distinguishes apoptosis from necrosis | Non-specific for apoptosis mechanism |
Disease Context Applications
Cancer Research Applications
In cancer research, apoptosis detection validates therapeutic efficacy and identifies resistance mechanisms [95].
Therapy Response Monitoring
Conventional therapies (chemotherapy, radiation) and targeted agents induce apoptosis through DNA damage and activation of intrinsic pathways. TEM analysis of tumor biopsies post-treatment reveals characteristic chromatin condensation and apoptotic bodies, confirming therapy-induced cell death [79]. Flow cytometric annexin V binding and caspase activation provide quantitative assessment of response kinetics.
Resistance Mechanism Elucidation
Dysregulated apoptosis enables cancer cell survival and therapeutic resistance [95]. Key resistance mechanisms include:
- BCL-2 family overexpression: Elevated BCL-2 blocks mitochondrial outer membrane permeabilization, preventing cytochrome c release [93] [66].
- IAP family upregulation: Inhibitor of Apoptosis Proteins directly bind and inhibit caspases [66].
- Death receptor pathway defects: Mutations in Fas/TNFR or downstream adaptors impair extrinsic apoptosis [79].
- p53 mutations: Loss of p53 function abrogates DNA damage-induced intrinsic pathway activation [79].
Multiparametric assessment combining TEM with BCL-2 immunohistochemistry, caspase activity assays, and death receptor expression profiling enables comprehensive resistance mechanism characterization.
Neurodegenerative Disease Applications
In neurodegenerative diseases, apoptosis contributes to selective neuronal loss, though the kinetics are typically slower than in cancer models [96].
Disease Mechanism Investigation
In Alzheimer's disease, amyloid-β and tau pathologies trigger neuronal apoptosis through mitochondrial dysfunction and oxidative stress. TEM reveals condensed chromatin and fragmented nuclei in vulnerable neuronal populations. Parkinson's disease involves α-synuclein accumulation and mitochondrial complex I impairment, activating intrinsic apoptosis. Caspase activation and cytochrome c release are detectable in substantia nigra neurons [96].
Therapeutic Development Assessment
Neuroprotective strategies aim to inhibit pathological apoptosis. TEM validates reduced ultrastructural apoptosis markers in preclinical models following treatment with caspase inhibitors, mitochondrial stabilizers, or neurotrophic factors. Annexin V imaging and TUNEL staining quantify neuroprotection efficacy in situ [96].
Integrated Experimental Workflows
A phased approach integrating multiple detection methods provides comprehensive apoptosis validation across research and drug development applications.
Figure 2: Integrated Apoptosis Detection Workflow. This diagram outlines a phased approach to apoptosis validation, progressing from early live-cell screening (blue) through biochemical confirmation (yellow) to morphological analysis (red), culminating in data integration (green).
Tiered Validation Strategy
- Tier 1: Early Screening: Implement live-cell compatible assays (annexin V, caspase probes, ΔΨm dyes) for kinetic analysis and high-throughput screening.
- Tier 2: Biochemical Confirmation: Apply fixed-cell methods (active caspase detection, western blotting for cleaved PARP, BCL-2 family profiling) to validate molecular mechanisms.
- Tier 3: Morphological Analysis: Utilize TEM and histology for ultrastructural confirmation and spatial context in tissues.
- Data Integration: Correlate findings across assays to establish conclusive evidence of apoptosis and exclude alternative cell death mechanisms.
Research Reagent Solutions
Table 2: Essential Reagents for Apoptosis Detection
| Reagent Category | Specific Examples | Primary Application | Key Features |
|---|---|---|---|
| Phosphatidylserine Detection | Annexin V-FITC, Annexin V-PE, Annexin V-BV421 | Flow cytometry, microscopy | Calcium-dependent binding, early marker, requires viability dye |
| Viability Probes | 7-AAD, Propidium Iodide, DRAQ7, Fixable Viability Stains | Flow cytometry, microscopy | Membrane integrity assessment, necrotic cell exclusion |
| Caspase Activity Detection | Fluorogenic substrates (DEVD-AMC, IETD-AFC), Cell-permeable probes (PhiPhiLux, NucView 488) | Spectrofluorometry, live-cell imaging | Specific cleavage sequences, kinetic measurements |
| Active Caspase Antibodies | Anti-active caspase-3 (PE conjugate), anti-cleaved PARP | Flow cytometry, western blot, IHC | Specific for activated forms, multiple application formats |
| Mitochondrial Probes | JC-1 (BD MitoScreen), TMRE, MitoTracker Red | Flow cytometry, microscopy | ΔΨm-sensitive accumulation, J-aggregate formation (JC-1) |
| DNA Fragmentation Kits | APO-BrdU TUNEL Assay, APO-DIRECT Kit | Flow cytometry, microscopy | End-labeling of DNA strand breaks, specific for late apoptosis |
| BCL-2 Family Antibodies | Anti-Bcl-2, Anti-Bax, Anti-Bad | Flow cytometry, western blot, IHC | Pathway mechanism elucidation, resistance marker detection |
Advanced Technical Considerations
Multiparametric Assay Design
Contemporary apoptosis research increasingly employs multiparametric approaches that simultaneously detect multiple events within individual cells. Eight-color flow cytometry panels can combine annexin V, viability dye, active caspase-3, BCL-2 family proteins, cell lineage markers, and cell cycle probes. This enables sophisticated analysis of apoptosis mechanisms in specific cellular subpopulations within heterogeneous samples, such as tumor microenvironments or mixed CNS cell cultures.
Quantification and Interpretation Guidelines
- Flow Cytometry: Establish gating strategies using untreated controls and inhibitor treatments (e.g., Z-VAD-FMK pan-caspase inhibitor). Include compensation controls for multicolor panels.
- TEM Morphometry: Apply quantitative stereology methods to determine apoptotic indices in tissue sections. Count at least 500 cells across multiple non-overlapping fields.
- Kinetic Analysis: For time-course experiments, collect samples at multiple timepoints (e.g., 0, 2, 4, 8, 16, 24 hours post-treatment) to capture apoptosis progression.
- Threshold Determination: Establish positive thresholds using isotype controls (antibodies), unstained cells (fluorophores), and inhibitor-treated samples.
Artifact Recognition and Troubleshooting
- Annexin V False Positives: Cell processing techniques (enzyme digestion, mechanical dissociation) can induce PS externalization. Use gentle handling and include appropriate controls.
- Caspase Assay Limitations: Some apoptotic pathways (e.g., caspase-independent) may not activate conventional caspase substrates. Confirm with complementary methods.
- TEM Artifacts: Improper fixation, dehydration, or sectioning can create morphological artifacts resembling apoptosis. Follow established protocols rigorously and include technical controls.
- Assay Interference: Test compounds (e.g., colored drugs, fluorescent molecules) may interfere with detection. Include compound controls without detection reagents.
This comprehensive technical guide provides researchers with validated methodologies for apoptosis detection and validation across disease contexts, with emphasis on integrating TEM with complementary approaches for definitive identification of early apoptotic events.
Transmission Electron Microscopy (TEM) has long been the gold standard for identifying the ultrastructural hallmarks of early apoptosis, including cytoplasmic shrinkage, chromatin condensation, and cavitation [65]. However, traditional TEM alone provides limited information about specific biochemical events. The emergence of Correlative Light and Electron Microscopy (CLEM) has revolutionized this field by combining the high-resolution structural capability of TEM with the molecular specificity of fluorescence microscopy and immunogold labeling [97] [98]. These integrated approaches allow researchers to precisely localize specific biomolecules within the context of detailed cellular ultrastructure, providing unprecedented insight into the early molecular events of apoptosis [99].
For apoptosis research, these techniques are particularly valuable for investigating early-stage events that precede morphological changes, such as phosphatidylserine externalization, caspase activation, and cytochrome c release [65] [100]. This technical guide explores the methodologies, applications, and quantitative approaches of correlative techniques within the context of early apoptosis research, providing researchers with practical frameworks for implementation.
Apoptosis Morphology and Detection: The Basis for Correlation
Ultrastructural Hallmarks of Apoptosis
Apoptosis progresses through distinct morphological phases that can be identified via TEM. Table 1 summarizes these stages and their key characteristics, which form the basis for correlative studies [65].
Table 1: Morphological Stages of Apoptosis Accessible via TEM
| Stage | Key Morphological Features | Detection Methods |
|---|---|---|
| Phase I (Early) | Cell shrinkage, decreased water content, increased eosinophilia, disappearance of microvilli, cavitation (vacuole formation) | TEM, fluorescence microscopy with membrane-permeant dyes |
| Phase IIa | Chromatin condensation (pyknosis), chromatin margination (assembly on inner nuclear membrane) | TEM, nuclear staining (Hoechst, DAPI), TUNEL assay |
| Phase IIb | Nuclear fragmentation, cytoskeleton degradation, membrane blebbing, apoptotic body formation | TEM, light microscopy (HE staining), Annexin V staining |
The diagram below illustrates the key morphological transitions during apoptosis that are detectable through electron microscopy:
Limitations of Single-Method Approaches
While TEM provides excellent spatial resolution for identifying apoptotic morphology, it cannot specifically label or identify the proteins and signaling molecules involved in apoptosis pathways [99]. Conversely, fluorescence microscopy reveals dynamic molecular events but lacks the resolution to place these events within precise subcellular contexts [98]. Correlative techniques bridge this gap by allowing researchers to first identify molecular events using fluorescence and then examine the underlying ultrastructure using TEM [97] [101].
Correlative Technique Methodologies
Immunogold Labeling for TEM
Immunogold labeling uses antibody-conjugated gold nanoparticles to mark specific antigens with high electron density, enabling their visualization via TEM. Table 2 outlines key reagent solutions and their functions in immunogold labeling protocols [99] [102].
Table 2: Essential Research Reagent Solutions for Immunogold Labeling
| Reagent/Material | Function | Application Notes |
|---|---|---|
| Primary Antibodies | Bind specifically to target antigens (e.g., caspases, cytochrome c) | Target apoptosis-specific markers; specificity must be validated |
| Gold-Conjugated Secondary Antibodies | Provide electron-dense tags for TEM visualization | Protein A gold or IgG gold commonly used; sizes typically 5-15 nm |
| FluoroNanogold (FNG) | Combinatorial probe containing both fluorophore and gold nanoparticle | Enables direct correlation between fluorescence and EM signals [98] |
| Saponin | Permeabilization agent | Preserves ultrastructure better than Triton X-100 [99] |
| Silver Enhancement Solutions | Catalytically deposit silver onto gold nanoparticles | Increases particle size from 1.4 nm to 20-30 nm for better visibility [97] |
Integrated CLEM Workflow for Apoptosis Research
The following diagram illustrates a generalized workflow for Correlative Light and Electron Microscopy, adapted for apoptosis studies:
Detailed Protocol: Immunogold Labeling for Apoptosis Markers
The following protocol is adapted from Fabig et al. for labeling ultrathin resin sections to detect apoptosis-related antigens [97]:
Sample Preparation
- Culture cells on gridded glass-bottom dishes (e.g., MatTek P35G-1.5-14-CGRD) to facilitate relocation [101]
- Induce apoptosis using appropriate stimuli (e.g., chemotherapeutic agents, UV irradiation)
- Fix cells with 4% formaldehyde with 0.1% glutaraldehyde in 0.1 M cacodylate buffer
Permeabilization and Blocking
- Permeabilize with 0.1% saponin for 10 minutes (preserves ultrastructure better than Triton X-100) [99]
- Block with 5% bovine serum albumin (BSA) for 30 minutes
Immunolabeling
- Incubate with primary antibodies against apoptosis markers (e.g., anti-cytochrome c, anti-caspase) for 1 hour
- Apply secondary detection:
- Option A: Simultaneous fluorescent (Alexa488/Alexa555) and immunogold labeling
- Option B: FluoroNanogold followed by silver enhancement [97]
Processing for TEM
- Post-fix with 2.5% glutaraldehyde
- Stain with 0.5% uranyl acetate and lead citrate
- Dehydrate through ethanol series and embed in epoxy resin (e.g., Epon or Lowicryl)
Imaging and Correlation
- First acquire fluorescence images to identify areas of interest
- Prepare ultrathin sections (70-90 nm) and collect TEM images of the same regions
- Correlate using grid coordinates and fiduciary markers
Advanced Applications in Apoptosis Research
Immunogold FIB-SEM for Volumetric Ultrastructural Analysis
Focused Ion Beam Scanning Electron Microscopy (FIB-SEM) combined with immunogold labeling enables 3D reconstruction of apoptotic cells at nanoscale resolution. This approach has been used to:
- Map the spatial distribution of epigenetic marks like H3K9me3 during neural stem cell apoptosis [99]
- Localize nuclear pore complexes in myoblasts differentiated on hydrogel surfaces [99]
- Reconstruct mitochondrial changes during early apoptosis in three dimensions
Quantitative Analysis of Immunogold Labeling
Advanced quantification methods enable researchers to determine the specific distribution of apoptotic markers within subcellular compartments:
Label Density Calculations
- LD(0): Raw gold particle count per unit area
- LD(-): Non-specific background labeling density
- LD(sp): Specific labeling density = LD(0) - LD(-) [102]
Improved Segmentation Parameters
- Standard deviation of pixel intensity combined with size and intensity parameters
- Enables differentiation of gold particles from similar-sized structures like ribosomes [102]
Background Subtraction Without Knockout/Knockdown
- Use regions outside the area of interest as background reference
- Verify with non-immune serum controls [102]
Technical Considerations and Optimization
Preservation of Ultrastructure and Antigenicity
Successful correlation depends on maintaining both structural preservation and antigen recognition:
- Fixation Optimization: Balance between formaldehyde concentration (preserves antigenicity) and glutaraldehyde concentration (preserves structure)
- Permeabilization Agent Selection: Saponin demonstrates superior ultrastructure preservation compared to Triton X-100, with significantly reduced nonspecific cytosolic immunolabeling (4% ± 2% vs. 38% ± 6%) [99]
- Embedding Media Compatibility: Lowicryl K4M and other methacrylates better preserve antigenicity for post-embedding labeling [97]
Correlation Accuracy and Workflow Efficiency
Several strategies enhance the precision and efficiency of correlation:
- Gridded Coverslips: Enable straightforward relocation of specific cells between imaging modalities [101]
- Fiducial Markers: Fluorescent beads or gold particles provide reference points for precise overlay
- Integrated Instruments: Newly developed systems combine light and electron microscopy in single instruments for simultaneous correlation [98]
The integration of TEM with fluorescence and immunogold labeling represents a powerful methodological advancement for apoptosis research. These correlative techniques enable researchers to connect specific molecular events with detailed ultrastructural changes occurring during early apoptosis, providing insights that were previously inaccessible through single-method approaches. As these technologies continue to evolve, particularly with improvements in volumetric imaging and quantitative analysis, they will undoubtedly yield deeper understanding of apoptotic pathways and facilitate development of novel therapeutic strategies for apoptosis-related diseases.
Conclusion
Transmission Electron Microscopy remains an indispensable tool for the definitive identification of early apoptosis, providing unmatched visualization of the key ultrastructural events that define this form of programmed cell death. While the technique demands specialized expertise and faces challenges in quantification, its role as a gold standard for morphological confirmation is unchallenged. The future of apoptosis research lies in integrated approaches that correlate TEM's detailed morphological insights with biochemical assays like caspase activation and phosphatidylserine exposure. As biomedical research advances, particularly in drug development and disease mechanism studies, TEM will continue to be crucial for validating the mode of action of novel therapeutics and understanding complex cell death pathways in human health and disease.
References
- 1. Apoptosis: A Review of Programmed Cell Death - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2117903/]
- 2. Apoptosis [https://en.wikipedia.org/wiki/Apoptosis]
- 3. Morphological assessment of apoptosis [https://www.sciencedirect.com/science/article/abs/pii/S1046202307002150]
- 4. A quantitative real-time approach for discriminating ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5253725/]
- 5. SEM vs TEM: Electron Microscopy Technique Comparison [https://measurlabs.com/blog/sem-vs-tem-comparison-of-electron-microscopy-techniques/]
- 6. Light Microscope vs. Electron Microscope [https://www.excedr.com/blog/light-microscope-vs-electron-microscope]
- 7. Recent Tumor Necrosis Factor-Related Apoptosis-Inducing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11922527/]
- 8. Targeting Cancer Cell Fate: Apoptosis, Autophagy, and ... [https://www.mdpi.com/1467-3045/47/6/460]
- 9. Understanding the Difference between Magnification and ... [https://www.nanoscience.com/blogs/understanding-the-difference-between-magnification-and-resolution-in-scanning-electron-microscopy/]
- 10. From mechanism to application: programmed cell death ... [https://www.sciencedirect.com/science/article/pii/S2452199X25002889]
- 11. Deep learning-driven automated mitochondrial segmentation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12125239/]
- 12. Key advantages - TEM [https://myscope.training/TEM_Key_advantages]
- 13. Detecting Apoptotic Human Lens Epithelial Cells With ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10599265/]
- 14. Programmed cell death detection methods: a systematic ... [https://link.springer.com/article/10.1007/s10495-022-01735-y]
- 15. High-Resolution Imaging of Morphological Changes ... [https://www.mdpi.com/2079-6374/15/8/522]
- 16. Light Microscopy as a Tool to Detect Apoptosis and Other ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12051451/]
- 17. The formation of the 'footprint of death' as a mechanism for ... [https://www.nature.com/articles/s41467-025-64206-3]
- 18. Tailoring of apoptotic bodies for diagnostic and therapeutic ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-024-05451-w]
- 19. Electronic detection of apoptotic cells on a microchip [https://www.sciencedirect.com/science/article/abs/pii/S0956566324007565]
- 20. Evaluating cell viability assessment techniques: a comparative ... [https://biomedical-engineering-online.biomedcentral.com/articles/10.1186/s12938-025-01452-y]
- 21. ultrastructural aspects of necrosis in human atherosclerosis [https://www.sciencedirect.com/science/article/abs/pii/S1054880723000443]
- 22. Morphological criteria to distinguish cell death induced by ... [https://link.springer.com/article/10.1007/s10495-005-6075-6]
- 23. Distinguishing between apoptosis and necrosis using a ... [https://www.sciencedirect.com/science/article/abs/pii/S0956566309000360]
- 24. Research review Screening and Detection of Apoptosis [https://www.sciencedirect.com/science/article/abs/pii/S0022480406004008]
- 25. Quantitative Analysis of Apoptosis and Necrosis in Live ... [https://pubmed.ncbi.nlm.nih.gov/36087259/]
- 26. analyzing ultrastructural features in cell death [https://pubmed.ncbi.nlm.nih.gov/39552095/]
- 27. Advancing Nanomedicine Through Electron Microscopy [https://pmc.ncbi.nlm.nih.gov/articles/PMC11899938/]
- 28. High-Resolution Imaging of Morphological Changes ... [https://pubmed.ncbi.nlm.nih.gov/40862983/]
- 29. Morphological and cytochemical determination of cell death ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2137940/]
- 30. Demystified …: Apoptosis - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC1186906/]
- 31. Morphologic criteria and detection of apoptosis [https://pubmed.ncbi.nlm.nih.gov/10412642/]
- 32. Cell Apoptosis/Methods for the detection of apoptosis [https://www.medchemexpress.com/literature/cell-apoptosis-methods-for-the-detection-of-apoptosis.html?srsltid=AfmBOorQPnmEg0f1K3F9r8oDmj2gOxOdpjlFioVckq6d36fT3MOfCG5p]
- 33. The essentials of developmental apoptosis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7047912/]
- 34. Distinction between Apoptosis and Oncosis | PLOS One [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0121674]
- 35. Immunoelectron microscopy: a comprehensive guide from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12420552/]
- 36. Sample Preparation of Adherent Cell Lines for Flow ... [https://pubmed.ncbi.nlm.nih.gov/39835228/]
- 37. Current trends in luminescence-based assessment of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10613953/]
- 38. Optimizing Apoptosis Assays: Scenario-Driven Guidance wit... [https://www.dihydro-b-erythroidine.com/index.php?g=Wap&m=Article&a=detail&id=32]
- 39. Brief Introduction to Contrasting for EM Sample Preparation [https://www.leica-microsystems.com/science-lab/life-science/brief-introduction-to-contrasting-for-em-sample-preparation/]
- 40. Early apoptosis induction in MCF-7 breast cancer cells ... [https://www.sciencedirect.com/science/article/abs/pii/S0141813025021567]
- 41. Hybrid silver–iron oxide nanoflowers: morphological tailoring ... [https://pubs.rsc.org/en/content/articlehtml/2025/ma/d5ma00210a]
- 42. Reynolds' Lead Citrate Stain [https://www.protocols.io/view/reynolds-39-lead-citrate-stain-cmnyu5fw]
- 43. Removal Of Lead Citrate and Uranyl Acetate Precipitates ... [https://www.emsdiasum.com/docstechnicaltechtipslead_citrate?srsltid=AfmBOoqIjytxaCLofEmQXn9OnqRGBxtahjfBv_xCJmxUjj5vtdOqsAYr]
- 44. High contrast en bloc staining of neuronal tissue for field ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3701260/]
- 45. High-contrast en bloc staining of neuronal tissue for field ... [https://www.nature.com/articles/nprot.2011.439]
- 46. Ultrastructural Complexity of Nuclear Components During ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4617388/]
- 47. Histopathological, ultrastructural, and biochemical traits of ... [https://www.sciencedirect.com/science/article/pii/S2590171022000583]
- 48. Ultrasound Targeted Apoptosis Imaging in Monitoring Early ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4101340/]
- 49. Molecular Imaging of Apoptosis: From Micro to Macro - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4377726/]
- 50. Research progress on morphology and mechanism of ... [https://www.nature.com/articles/s41419-024-06712-8]
- 51. Toxoplasma gondii Inhibits Apoptosis in Infected Cells by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2687828/]
- 52. Imaging of morphological and biochemical hallmarks ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6851291/]
- 53. Evaluation of image analysis tools for the measurement ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2025.1572212/full]
- 54. Community-developed checklists for publishing images ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10922596/]
- 55. Detecting Apoptotic Human Lens Epithelial Cells With ... [https://pubmed.ncbi.nlm.nih.gov/37885524/]
- 56. Optimization of protocols for pre-embedding immunogold ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8173732/]
- 57. Quantitative spatial analysis of crystallin proteins in human ... [https://www.nature.com/articles/s41598-025-17896-0]
- 58. Optimization of negative stage bias potential for faster ... [https://www.sciencedirect.com/science/article/pii/S2590152421000039]
- 59. Transformer-based spatial-temporal detection of apoptotic ... [https://elifesciences.org/reviewed-preprints/90502]
- 60. 3D correlative light and electron microscopy reveals the ... [https://www.sciencedirect.com/science/article/pii/S1549963425000735]
- 61. Time‐Dependent Effects of Low‐Intensity Pulsed Ultrasound ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12191408/]
- 62. Cells die with increased cytosolic ATP during apoptosis [https://www.nature.com/articles/4401661]
- 63. Researchers identify new molecular switch involved in ... [https://www.news-medical.net/news/20251103/Researchers-identify-new-molecular-switch-involved-in-programmed-cell-death.aspx]
- 64. Insights on the crosstalk among different cell death ... [https://www.nature.com/articles/s41420-025-02328-9]
- 65. Apoptosis detection: a purpose-dependent approach ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8208110/]
- 66. Programmed cell death detection methods - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC9308588/]
- 67. Guidelines for Regulated Cell Death Assays - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC7970050/]
- 68. Comparative Analysis of Autophagy and Apoptosis in Disc ... [https://www.mdpi.com/1422-0067/25/4/2352]
- 69. Quantitative assessment of cell fate decision between ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5730598/]
- 70. Apoptosis western blot guide [https://www.abcam.com/en-us/knowledge-center/western-blot/western-blot-analysis-of-apoptosis]
- 71. Cell Apoptosis/Methods for the detection of apoptosis [https://www.medchemexpress.com/literature/cell-apoptosis-methods-for-the-detection-of-apoptosis.html?srsltid=AfmBOoqiWVxilHWPV6L06hpm97D7aItO7i8nW2xcvSiSAUopqsdY8ZJj]
- 72. Protocol for specific induction of apoptosis or necroptosis in ... [https://pubmed.ncbi.nlm.nih.gov/41205180/]
- 73. Protocol for specific induction of apoptosis or necroptosis in ... [https://www.sciencedirect.com/science/article/pii/S266616672500591X]
- 74. Automated detection of apoptotic bodies and cells in label-free ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10563152/]
- 75. Emerging Electrochemical Approaches for the Early Detection ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12355238/]
- 76. TEM Analysis - Semiconductor [https://www.thermofisher.com/blog/semiconductors/tem-analysis-semiconductor-development/]
- 77. Optimizing SEM Parameters - Purdue College of Agriculture [https://ag.purdue.edu/department/arge/facilities-and-research/microscopy/training/optimizing-sem-parameters.html]
- 78. Three Distinct Stages of Apoptotic Nuclear Condensation ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2705844/]
- 79. Detection of apoptosis : A review of conventional and novel... [https://pubs.rsc.org/en/content/articlehtml/2010/ay/c0ay00247j]
- 80. Cell Apoptosis/Methods for the detection of apoptosis [https://www.medchemexpress.com/literature/cell-apoptosis-methods-for-the-detection-of-apoptosis.html?srsltid=AfmBOopOgmZRtvWr6_we7ILOdIXAYXkKYeYBJ7GPg3De2gsWy4iGZ28l]
- 81. Study of caspase-6 activity in aggressive HCT116 cells using ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12214825/]
- 82. Thapsigargin triggers a non-apoptotic, caspase ... [https://www.nature.com/articles/s41420-025-02602-w]
- 83. Assay Kit in the Real World: 5 Uses You'll Actually See... Apoptosis [https://www.linkedin.com/pulse/apoptosis-assay-kit-real-world-5-uses-youll-actually-sry9f]
- 84. Targeting regulated cell death: Apoptosis, necroptosis ... [https://pubmed.ncbi.nlm.nih.gov/40115032]
- 85. Assessment of apoptosis and autophagy in oral squamous ... [https://link.springer.com/article/10.1007/s10266-025-01236-9]
- 86. "Reading cell death with light: Real-time visualization of ... [https://www.eurekalert.org/news-releases/1096590]
- 87. Apoptosis induction capability of silver nanoparticles ... [https://www.sciencedirect.com/science/article/pii/S2405844024004316]
- 88. Cell Apoptosis/Methods for the detection of apoptosis [https://www.medchemexpress.com/literature/cell-apoptosis-methods-for-the-detection-of-apoptosis.html?srsltid=AfmBOor-LuE_78gbQL3MKfmIinVQdPfN2M9Mf9lHj072LaXidANMzW3z]
- 89. Guidelines for Regulated Cell Death Assays: A Systematic ... [https://www.frontiersin.org/journals/cell-and-developmental-biology/articles/10.3389/fcell.2021.634690/full]
- 90. Call to action to properly utilize electron microscopy ... [https://www.sciencedirect.com/science/article/pii/S0171933523000808]
- 91. Targeting ER stress-mediated apoptosis by MSC-derived ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12554144/]
- 92. MODULATION OF APOPTOTIC PATHWAYS AND ... [https://www.sciencedirect.com/science/article/pii/S2667031325001940]
- 93. Detection | Apoptosis Signaling Apoptosis [https://www.bdbiosciences.com/en-sg/learn/research/cell-biology/apoptosis]
- 94. Enhancing Contrast, Adding Scale Bars to TEM Images [https://www.nanoimagingservices.com/about/blog/enhancing-contrast-and-adding-scale-bars-to-tem-images]
- 95. Cell Death: A New Frontier in Cancer Research [https://www.frontiersin.org/research-topics/70340/cell-death-a-new-frontier-in-cancer-researchundefined]
- 96. Precision medicine in neurodegenerative diseases [https://www.sciencedirect.com/science/article/abs/pii/S0079612325001232]
- 97. Labeling of ultrathin resin sections for correlative light and ... [https://pubmed.ncbi.nlm.nih.gov/22857924/]
- 98. Correlative Fluorescence and Electron Microscopy - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4211606/]
- 99. Immunogold FIB-SEM: Combining Volumetric Ultrastructure ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6778054/]
- 100. Current trends in luminescence-based assessment of apoptosis [https://pubs.rsc.org/en/content/articlehtml/2023/ra/d3ra05809c]
- 101. Some tips and tricks for a Correlative Light Electron ... [https://www.sciencedirect.com/science/article/abs/pii/S0091679X24000633]
- 102. Improved quantification of immune-gold labeling and its ... [https://www.sciencedirect.com/science/article/abs/pii/S0968432821000512]